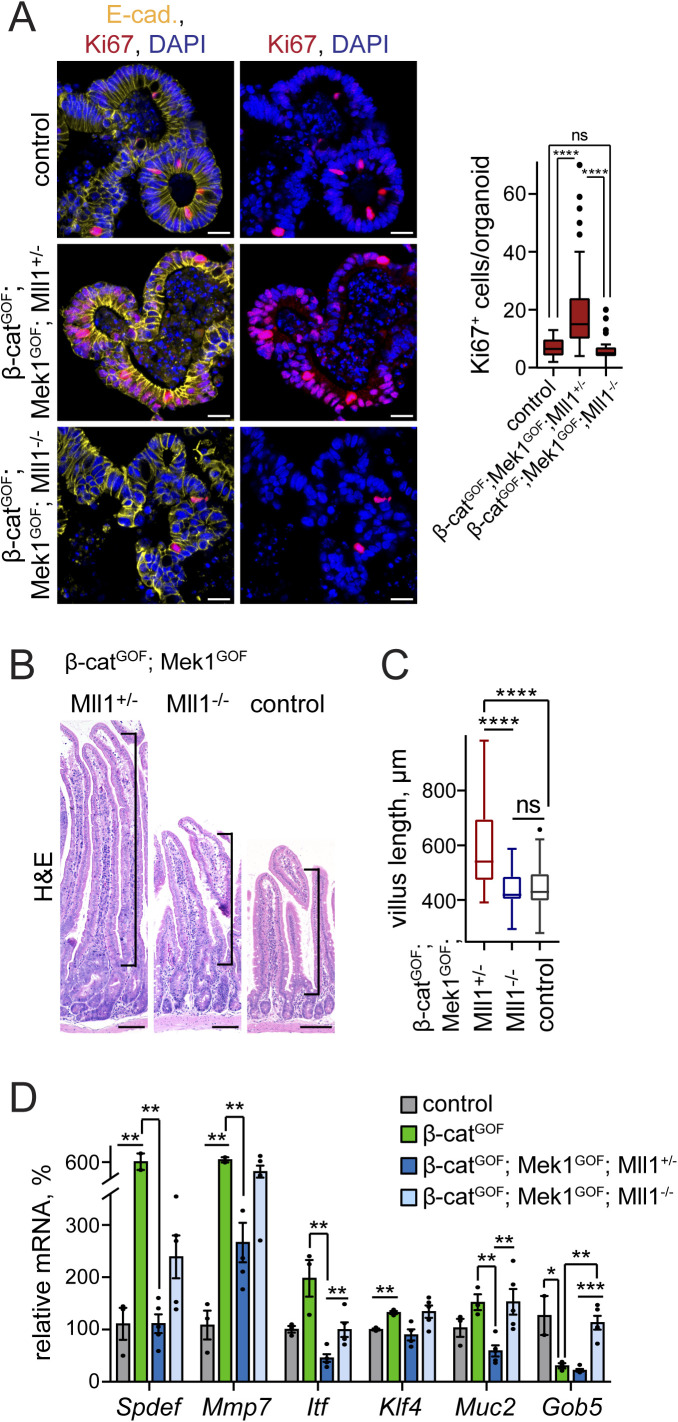
Figure S5. Loss of Mll1 abolishes proliferation and promotes Paneth/goblet cell differentiation in intestinal organoids and crypts with concomitant activation of Wnt and Mapk signalling.
(A) Immunofluorescence staining for Ki67 (red) on sections of non-induced control and tamoxifen-induced β-catGOF; Mek1GOF; Mll1+/− and β-catGOF; Mek1GOF; Mll1−/− organoids. E-cadherin (yellow) stains cell borders, nuclei in blue (DAPI), scale bars 20 μm. Right: Quantification of Ki67+ cells per organoid in control, β-catGOF; Mek1GOF; Mll1+/− and β-catGOF; Mek1GOF; Mll1−/− organoids, n = 4 independent experiments, Mann–Whitney U test, ****P < 0.0001. Box plot indicates median (middle line) and 25th, 75th percentile (box) with Tukey whiskers. (B) H&E staining on sections of small intestines of β-catGOF; Mek1GOF; Mll1+/− and β-catGOF; Mek1GOF; Mll1−/− at 20 d after induction with tamoxifen and wild-type control mice, scale bars 100 μm. Stainings were performed in n = 3 independent wild-type and n = 5 independent mutant mice per genotype. (C) Quantification of villus length in n = 5 independent β-catGOF; Mek1GOF; Mll1+/− and β-catGOF; Mek1GOF; Mll1−/− mice each at 20 d after induction relative to n = 3 independent control mice, at least 10 villi per mouse measured, Mann–Whitney U test, ****P < 0.0001. Box plot indicates median (middle line) and 25th, 75th percentile (box) with Tukey whiskers. (D) mRNA expression of secretory lineage markers Spdef, Mmp7, Itf, Klf4, Muc2, and Gob5 in intestines of control (grey, n = 3 independent mice), β-catGOF (green, n = 3 independent mice), β-catGOF; Mek1GOF; Mll1+/− (blue, n = 5 independent mice), and β-catGOF; Mek1GOF; Mll1−/− mice (light blue, n = 5 independent mice) at 20 d after the induction of mutagenesis, two-tailed unpaired t test, Spdef: **P = 0.009 (control/β-catGOF), **P = 0.0046 (β-catGOF/Mll1+/−), Mmp7: **P = 0.0012 (control/β-catGOF), **P = 0.0026 (β-catGOF/Mll1+/−), Itf: **P = 0.0014 (β-catGOF/Mll1+/−), **P = 0.008 (Mll1+/−/Mll1−/−), Klf4: **P = 0.009, Muc2: **P = 0.002 (β-catGOF/Mll1+/−), **P = 0.007 (Mll1+/−/Mll1−/−), Gob5: *P = 0.04, **P = 0.003, ***P = 0.00015. Data are presented as mean values ± SEM.