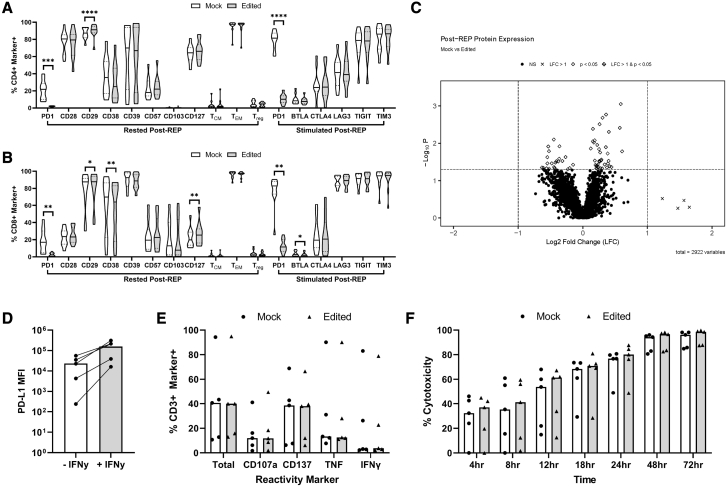
Figure 4.
PD-1-targeting CRISPR-Cas9 has minimal effects on expanded TIL functionality and phenotype
(A) Flow cytometry phenotype analysis of mock (white) and edited (gray) CD4+ REP-TILs post-REP. Samples were rested and stained or stimulated for 48 h with aCD3/aCD28 beads prior to staining. Markers analyzed can be found on the x axis. TEM, effector memory phenotype; TCM, central memory phenotype; Treg, regulatory phenotype. Solid and dotted lines represent median and quartiles, respectively. The average of two replicates was used for each of the 10 samples. Statistical significance was tested via paired t tests or Wilcoxon matched-pairs signed rank test ((∗p ≤ 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). (B) Same as A but for CD8+ REP-TILs. (C) Volcano plot comparing expression of 2,922 proteins from post-REP mock and edited REP-TILs. Each dot represents average expression (10 samples per condition, 2 replicates per sample) of one identified protein. Crosses signify proteins upregulated (log2 fold change >1) in mock (positive values) or edited (negative values), and diamonds denote statistically significant effects (paired t test, p ≤ 0.05). Differentially expressed proteins match both criteria and are shown as cross within diamond. (D) PD-L1 MFI of autologous tumor cell lines used for reactivity and cytotoxicity experiments, with and without IFNγ treatment. All reactivity and cytotoxicity tests described were carried out with IFNγ-treated cells. (E) Upregulation of reactivity markers in mock and edited CD3+ TILs measured via flow cytometry after 8-h co-culture with PD-L1 expressing autologous tumor cell lines. An average of three replicates was used for each of the five samples. Bars signify median. (F) Cytotoxicity of mock and edited cells during 72-h co-culture with PD-L1 expressing autologous tumor cell lines. Cytotoxicity calculated relative to tumor alone (negative) and cytotoxic agent (positive) controls. An average of three replicates was used for each of the five samples. Bars signify median. (A, B, E, and F) White bars and shaded bars represent mock and edited samples, respectively. (D–F) Experiments were carried out using three MM, one ovarian, and one sarcoma sample (see Materials and methods for details).