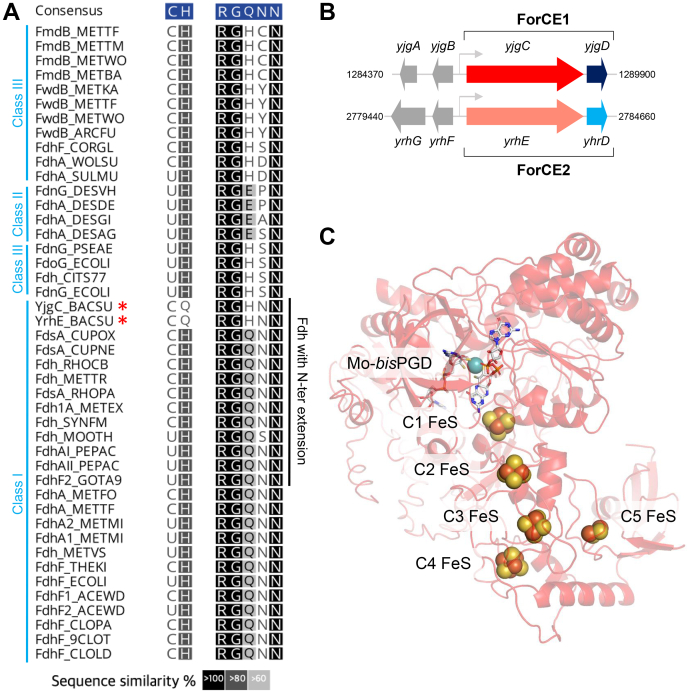
Figure 1.
Bacillus subtilis genome encodes putative noncanonical FDHs.A, alignment of the amino acids involved in the active site of characterized FDH enzymes and the two putative FDH proteins from B. subtilis (red asterisks). Consensus sequence (blue box) shows which amino acids are conserved with a 50% threshold. FDHs have been organized into three classes according to the sequence and organization of their active site (Fig. S7): Class I: C/UH RGQ or C/UQ RGH, class II: C/UH RGE, and class III: C/UH RGH. Overview of the full sequence alignment is shown in Figure S1B. B, genomic organization of yjgCD (ForCE1) and yrhED (ForCE2) in B. subtilis. C, structural model of YjgC generated with I-TASSER (89) using FdsA from R. capsulatus (PDB ID: 6TGA). FeS clusters are named according to their proximity to the Mo cofactor in the 3D structural model. FDHs, formate dehydrogenases.