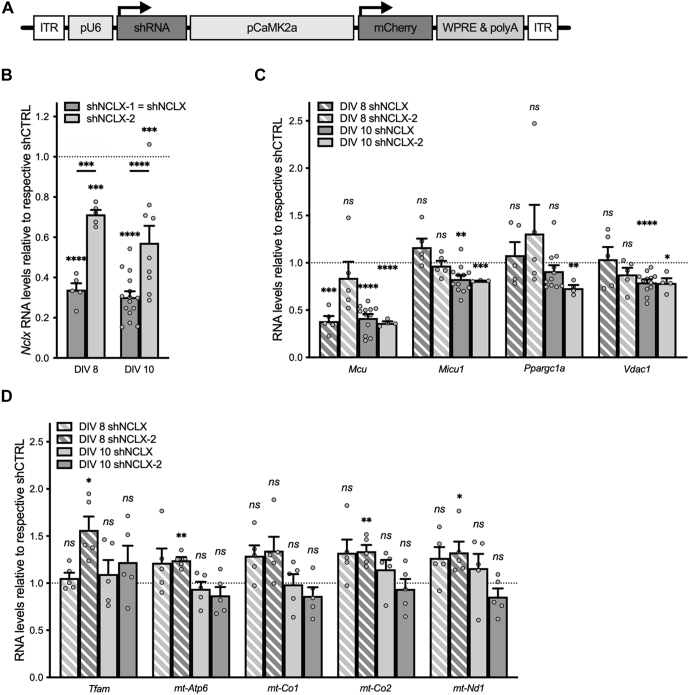
Figure 1.
shRNA-mediated knockdown of NCLX in primary hippocampal cultures.A, rAAV construct design. Primary hippocampal cultures were infected on DIV 3 with rAAVs driving expression of two different shRNAs directed against Nclx (shNCLX-1 and shNCLX-2; shNCLX-1 is hereafter referred to as shNCLX) or a control shRNA with no known targets in the mouse genome (shCTRL) under control of the U6 promoter (pU6), with mCherry as a marker protein expressed under control of the CaMK2a promoter (pCaMK2a). B, quantitative RT–PCR (qRT–PCR) analysis of the Nclx message (normalized to Gusb and expressed as a fraction of the levels in rAAV-shCTRL-infected cultures) on 5 and 7 days after infection, on DIV 8 and DIV 10 (n = 3–15 independent cultures; two-tailed one-sample t tests versus a hypothetical value of one; DIV 8: shNCLX-1: t(4) = 20.80, p < 0.0001, shNCLX-2: t(4) = 12.77, p = 0.0002. DIV 10: shNCLX-1: t(14) = 23.937, p < 0.0001, shNCLX-2: t(8) = 5.07, p = 0.0010; mixed-effects model one-way ANOVA followed by Šidák's multiple comparisons test; shNCLX versus shNCLX-2: DIV 8 t(5,5) = 5.717, p = 0.0001, DIV 10: t(15,9) = 5.762, p < 0.0001). C, qRT–PCR analysis of two genes involved in mitochondrial calcium signaling (Mcu and Micu1), the major regulator of mitochondrial biogenesis (Ppargc1a), and Vdac1 (all normalized to Gusb and expressed as a fraction of the levels observed in rAAV-shCTRL-infected sister cultures) 5 and 7 days after infection, on DIV 8 and DIV 10 (n = 3–12 independent cultures; two-tailed one-sample t tests versus a hypothetical value of one; Mcu: DIV 8 shNCLX: t(4) = 11.39, p = 0.0003, DIV 8 shNCLX-2: t(4) = 0.9359, p = 0.4023, DIV 10 shNCLX: t(11) = 14.10, p < 0.0001, DIV 10 shNCLX-2: t(3) = 38.43, p < 0.0001; Micu1: DIV 8 shNCLX: t(4) = 1.846, p = 0.1387, DIV 8 shNCLX-2: t(4) = 0.5948, p = 0.5840, DIV 10 shNCLX: t(11) = 4.157, p = 0.0016, DIV 10 shNCLX-2: t(3) = 4.549, p = 0.0199; Ppargc1a: DIV 8 shNCLX: t(4) = 0.5793, p = 0.5934, DIV 8 shNCLX-2: t(10) = 1.021, p = 0.3651, DIV 10 shNCLX: t(1) = 1.397, p = 0.1926, DIV 10 shNCLX-2: t(3) = 8.033, p = 0.0040; Vdac1: DIV 8 shNCLX: t(4) = 0.2977, p = 0.7807, DIV 8 shNCLX-2: t(4) = 1.752, p = 0.1547, DIV 10 shNCLX: t(11) = 6.436, p < 0.0001, DIV 10 shNCLX-2: t(3) = 4.286, p = 0.0233). D, qRT–PCR analysis of the mitochondrial transcription factor Tfam and four mitochondrial genes that encode members of the electron transport chain (n = 5 independent cultures; two-tailed one-sample t tests versus a hypothetical value of one; Tfam: DIV 8 shNCLX: t(4) = 0.9556, p = 0.3934, DIV 8 shNCLX-2: t(4) = 3.976, p = 0.0165, DIV 10 shNCLX: t(4) = 0.6539, p = 0.5489, DIV 10 shNCLX-2: t(4) = 1.286, p = 0.2679; mt-Atp6: DIV 8 shNCLX: t(4) = 1.442, p = 0.2227, DIV 8 shNCLX-2: t(4) = 7.184, p = 0.0020, DIV 10 shNCLX: t(4) = 0.7996, p = 0.4687, DIV 10 shNCLX-2: t(4) = 1.463, p = 0.2173; mt-Co1: DIV 8 shNCLX: t(4) = 2.638, p = 0.0577, DIV 8 shNCLX-2: t(4) = 2.340, p = 0.0794, DIV 10 shNCLX: t(4) = 0.1198, p = 0.9104, DIV 10 shNCLX-2: t(4) = 1.480, p = 0.2131; mt-Co2: DIV 8 shNCLX: t(4) = 2.328, p = 0.0804, DIV 8 shNCLX-2: t(4) = 5.174, p = 0.0066, DIV 10 shNCLX: t(4) = 1.491, p = 0.2102, DIV 10 shNCLX-2: t(4) = 0.5774, p = 0.5946; mt-Nd1: DIV 8 shNCLX: t(4) = 2.349, p = 0.0786, DIV 8 shNCLX-2: t(4) = 2.848, p = 0.0465, DIV 10 shNCLX: t(4) = 1.063, p = 0.3477, DIV 10 shNCLX-2: t(4) = 1.656, p = 0.1730). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Bar graphs show the mean + SEM. DIV, day in vitro; ITR, inverted terminal repeat; Mcu, mitochondrial uniporter; Micu1, mitochondrial calcium uptake 1; mt-Atp6, ATP synthase F0 subunit 6, mitochondrial; mt-Co1, cytochrome c oxidase I, mitochondrial; mt-Co2, cytochrone c oxidase II, mitochondrial; mt-Nd1, NADH dehydrtogenase I, mitochondrial; NCLX, solute carrier family 8 sodium/calcium/lithium exchanger, member B1; ns, not significant; pCaMK2a, calcium/calmodulin-dependent protein kinase II alpha promoter; Ppargc1a, peroxisome proliferative activated receptor, gamma, coactivator 1 alpha; pU6, U6 small nuclear RNA promoter; qRT–PCR, quantitative reverse transcription polymerase chain reaction; rAAV, recombinant adeno-associated viral vector; shRNA, short hairpin RNA; Tfam, transcription factor A, mitochondrial; Vdac1, voltage-dependent anion channel 1; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.