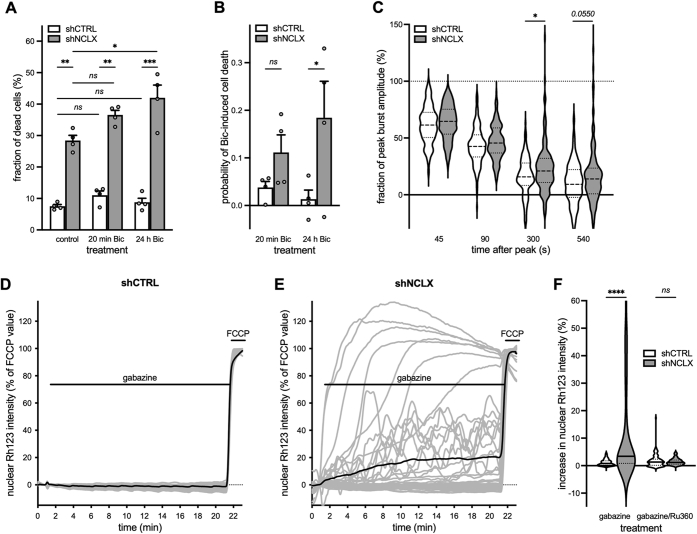
Figure 6.
shRNA-mediated knockdown of NCLX impairs mitochondrial calcium extrusion following action potential bursts, compromises ΔΨmduring neuronal activity, and renders synaptic activity neurotoxic.A and B, primary hippocampal cultures infected with either rAAV-shCTRL or rAAV-shNCLX were stimulated with the GABAAR antagonist, bicuculline (Bic; 50 μM) for 20 min or 24 h to evoke action potential bursting, and then the numbers of live and dead cells assessed after 24 h (n = 4 independent cultures for all conditions). A, proportions of dead cells (repeated-measures two-way ANOVA followed by Tukey's multiple comparisons test; main effect of shRNA F(1,3) = 283.0, p = 0.0005; shCTRL versus shNCLX: control q(4,4) = 9.219, p = 0.0048, 20 min Bic q(4,4) = 11.27, p = 0.0016, 24 h Bic q(4,4) = 14.66, p = 0.0004; main effect of treatment F(2,6) = 6.042, p = 0.0365; control versus 20 min Bic: shCTRL q(4,4) = 1.543, p = 0.8692, shNCLX q(4,4) = 3.590, p = 0.2454; control versus 24 h Bic: shCTRL q(4,4) = 0.5523, p = 0.9982, and shNCLX q(4,4) = 5.990, p = 0.0383). B, quantification of the probability of cells dying because of Bic treatment, which takes into account elevated levels of basal cell death (see the Experimental procedures section; repeated-measures two-way ANOVA followed by Šídák's multiple comparisons test; shNCLX versus shCTRL: 20 min t(4,4) = 2.493, p = 0.1688, 24 h t(4,4) = 5.807, p = 0.0202). C, the GABAAR antagonist, gabazine (5 μM), was used to trigger a single action potential burst in primary hippocampal cultures coinfected with rAAVs driving expression of the mitochondrially targeted FRET-based calcium indicator 4mtD3cpv under control of the CamK2a promoter and either shCTRL or shNCLX. Shown are mitochondrial calcium levels as measured 45, 90, 300, and 540 s after the peak response and normalized to the peak amplitude (shCTRL n = 159 cells from three coverslips and three preparations; shNCLX n = 158 cells from three coverslips and three preparations; Kruskal–Wallis test followed by Dunn's multiple comparisons test; shCTRL versus shNCLX: 45 s Z(159,158) = 0.6242, p = 0.5325, 90 s Z(159,158) = 1.186, p = 0.2356, 300 s Z(159,158) = 2.129, p = 0.0333, and 540 s Z(159,158) = 1.919, p = 0.0550). D–F, gabazine (5 μM) was applied in the presence (or not) of the MCU antagonist Ru360 (10 μM) to primary hippocampal cultures infected with rAAVs driving the expression of either shNCLX or shCTRL and loaded with Rh123. When used, Ru360 was present in the culture medium ≥30 min prior to and during the entire course of the experiment. The mitochondrial uncoupler FCCP (5 μM) was used to trigger complete ΔΨm breakdown. D and E, representative changes in nuclear Rh123 fluorescence changes to gabazine stimulation in rAAV-shCTRL-infected (D) and rAAV-shNCLX-infected cells (E) on a single coverslip each (gray, individual cells; black, their mean). F, quantification of changes in ΔΨm (gabazine: shCTRL n = 194 cells from six coverslips and five preparations, shNCLX n = 171 cells from four coverslips and three preparations; gabazine/Ru360: shCTRL n = 121 cells from three coverslips and three preparations, shNCLX n = 132 cells from four coverslips and three preparations), expressed as the peak amplitude of the nuclear Rh123 intensity during 20 min gabazine stimulation (Kruskal–Wallis test followed by Dunn's multiple comparisons test; shCTRL versus shNCLX: gabazine Z(194,171) = 8.770, p < 0.0001, gabazine + Ru360 Z(121,132) = 0.5102, p = 0.6099). ns, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Bar graphs show the mean + SEM. Violin plots show the probability density of the data as well as median and quartile divisions. Bic, bicuculline; CamK2a, calcium/calmodulin dependent protein kinase II alpha; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; FRET, Förster resonance energy transfer; GABAAR, gamma-aminobutyric acid (GABA) A receptor; MCU, mitochondrial calcium uniporter; NCLX, solute carrier family 8 sodium/calcium/lithium exchanger, member B1; ns, not significant; rAAV, recombinant adeno-associated viral vector; Rh123, rhodamine 123; Ru360, ruthenium 360; ΔΨm, mitochondrial membrane potential.