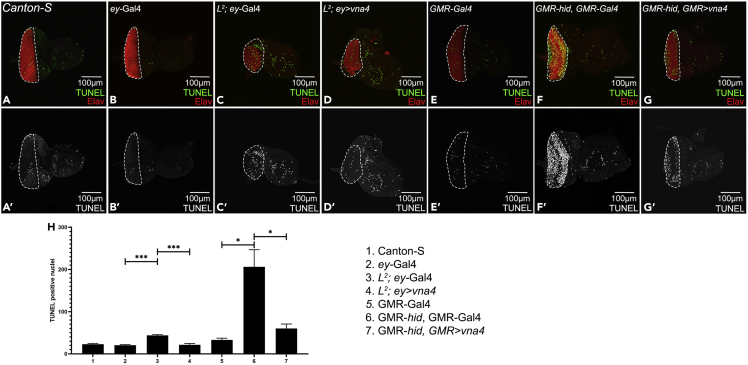
Figure 2.
TUNEL assay labels free 3′-OH DNA ends
(A–G) Eye-antennal imaginal discs stained for Elav (red) and showing TUNEL positive nuclei (green), and (A′–G′) split channel for TUNEL in (A, A′) Canton-S, (B, B′) ey-Gal4, (C, C′) L2; ey-Gal4, (D, D′) L2; ey>vna4, (E, E′) GMR-Gal4, (F, F′) GMR-hid, GMR-Gal4, and (G, G′) GMR-hid, GMR>vna4. (H) Quantification of the TUNEL positive nuclei (green) in the fly retina (marked by dotted line) shows that the average number of dying cells were significantly reduced when vna4, a newt gene is misexpressed in the GMR-hid, GMR-Gal4 or L2-mutant background as compared to GMR-hid, GMR-Gal4 or L2-mutant only. The orientation of all imaginal discs is dorsal up and posterior left. The magnification of all discs is 20×. A total of five eye-antennal imaginal discs (n = 5) was analyzed for each genotype. Error bars show standard deviation (mean ± SEM), and symbols above the error bar signify as ∗∗∗ p-value <0.001, ∗∗ p-value <0.01, ∗ p-value <0.05, and ns, not significant, p-value >0.05 respectively.