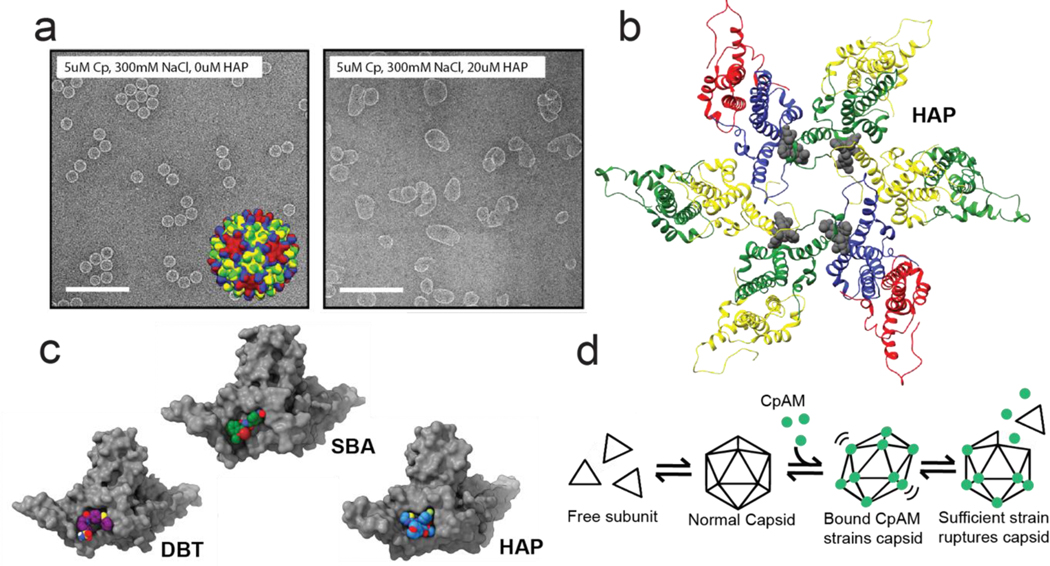
Figure 2. The effects of CpAM binding to HBV core protein.
(a) Electron micrographs of assembled HBV Cp dimer. (left panel) Virus-like particles are ~35nm icosahedra. Inset: an model of a T=4 capsid structure with chains colored according to quasi-equivalent environment. When assembled in the presence of a HAP (right panel), reaction products include large misassembled structures (40). (b) A hexamer fragment of a T=4 capsid structure in complex with a HAP (44). Coloring is the same as in the inset in the left panel of a, except with the HAP molecules colored gray. The HAP molecules bind at the interface between two adjacent subunits. (c) One site, multiple chemistries: a selection of CpAMs which target the interface between HBV capsid protein subunits. Despite occupying the same site, each molecule has a distinct binding mode, and associated phenotype (47, 49, ••52) (d) A simplified reaction schematic for how CpAMs disrupt pre-formed capsids. Increasing the per-contact association energy causes favorable, but modified pairwise orientation that induces global strain. Strained capsids will rupture to relieve strain (53). Components in this figure are extracted from or adapted from figures in references (40), (44), and (••52).