Figure 1.
Simultaneous analysis of HIV-1 DNA sequence, integration site, and transcriptional activity from individual infected cells
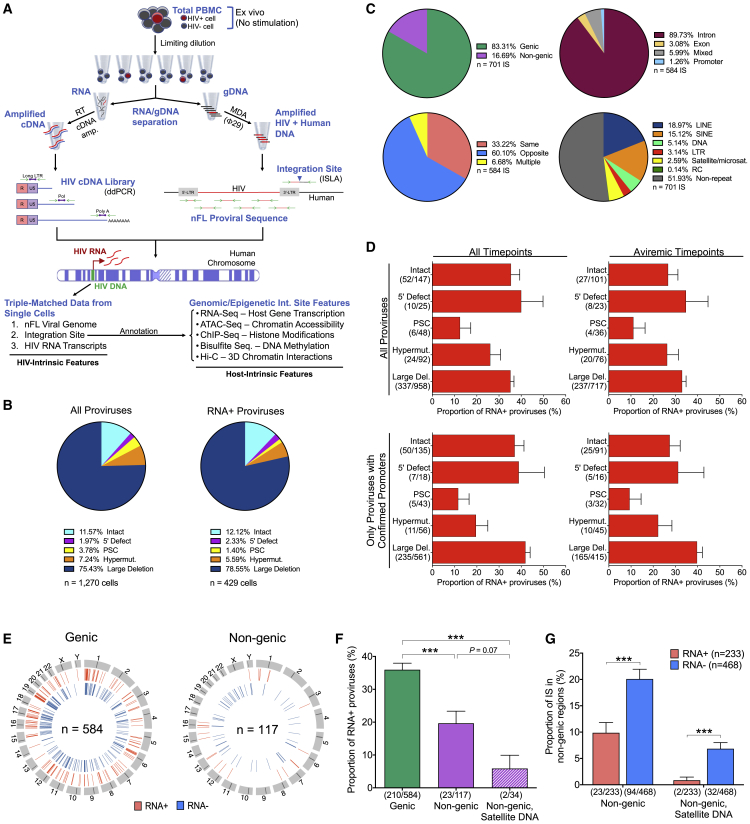
(A) Schematic representation of the PRIP-seq assay design.
(B) Proviral sequence classification in all analyzed HIV-1-infected cells and in long LTR RNA-expressing HIV-1-infected cells (PSC, premature stop codon).
(C) Proportions of proviruses in genic versus nongenic positions, introns/exons/promoters (genic sites only), same or opposite orientation to host genes (genic sites only), and repetitive genomic elements.
(D) Proportion of HIV-1 long LTR RNA-expressing proviruses among analyzed proviruses, stratified according to proviral sequence intactness/defects. Data are shown separately for all proviruses, all proviruses collected during aviremic time points, and for a subset of proviruses with experimentally confirmed intact core promoter regions.
(E) Circos plots reflecting the chromosomal locations of transcriptionally active (RNA+) and silent (RNA−) proviruses in genic versus nongenic DNA.
(F) Proportion of transcriptionally active proviruses among proviruses integrated in either genic, nongenic, or nongenic satellite DNA regions.
(G) Contribution of proviruses in nongenic or nongenic satellite DNA to the total number of transcriptionally active (RNA+) or silent proviruses (RNA−) with detectable chromosomal IS.
(E–G) HIV-1 long LTR RNA-expressing proviruses were considered “RNA+.” (∗∗∗p < 0.001, Fisher’s exact tests were used for all comparisons. Error bars represent standard errors of proportions ).