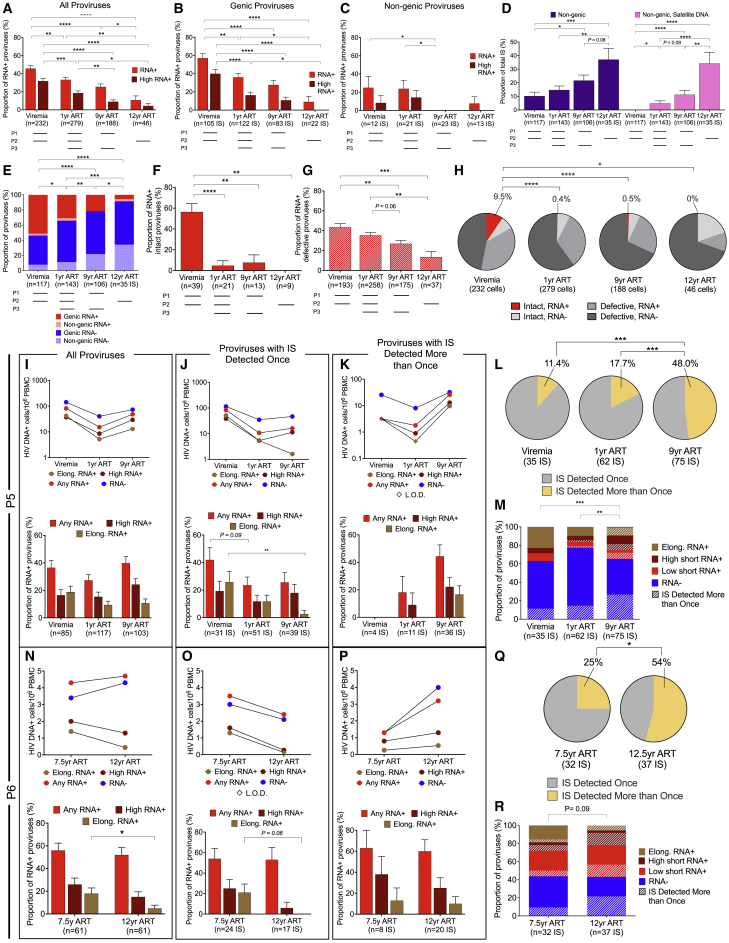
Figure 3.
Longitudinal evolution of HIV-1 proviruses
(A–C) Relative proportions of proviruses expressing any HIV-1 RNA or high-level (>10,000 postamplification copies) HIV-1 RNA at indicated time points for participants 1–3 (P1–P3). Data for all proviruses (A), proviruses integrated in genic locations (B), and proviruses in nongenic locations (C) are shown; (B) and (C) only include proviruses for which IS are available.
(D) Proportion of proviruses integrated in nongenic and nongenic, satellite DNA in a combined longitudinal analysis of participants 1–3.
(E) Relative contribution of RNA-positive or -negative proviruses in genic versus nongenic chromosomal locations to the total number of proviruses with known IS in P1–P3.
(F and G) Proportions of intact (F) and defective (G) proviruses that were transcriptionally active in P1–P3 at indicated longitudinal time points.
(H) Contribution of indicated proviruses to the total number of proviruses in participants 1–3.
(A–G) Horizontal dashes indicate available time points from each participant; HIV-1 long LTR RNA-expressing proviruses were considered “RNA+.”
(I–K and N–P) Frequencies and proportions of proviruses expressing any HIV-1 RNA, high-level (>10,000 postamplification copies) HIV-1 RNA, elongated HIV-1 RNA (containing pol, nef, spliced tat-rev, or poly-A sequences), or no HIV-1 RNA in study participants 5 (P5, I–K) and 6 (P6, N–P). Data for all proviruses (I and N), for proviruses with IS detected once (J and O), and for proviruses with IS detected more than once (K and P) are shown.
(J, K, O, and P) Only include proviruses for which IS are available.
(L and Q) Contribution of proviruses with IS detected once or multiple times to the total number of proviruses with known IS in participants 5 (L) and 6 (Q). In (M/R), proviruses are additionally stratified by HIV-1 RNA expression status. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Mann-Whitney U tests, Fisher’s exact tests, or G tests were used for all comparisons. Error bars in bar diagrams represent SEP).