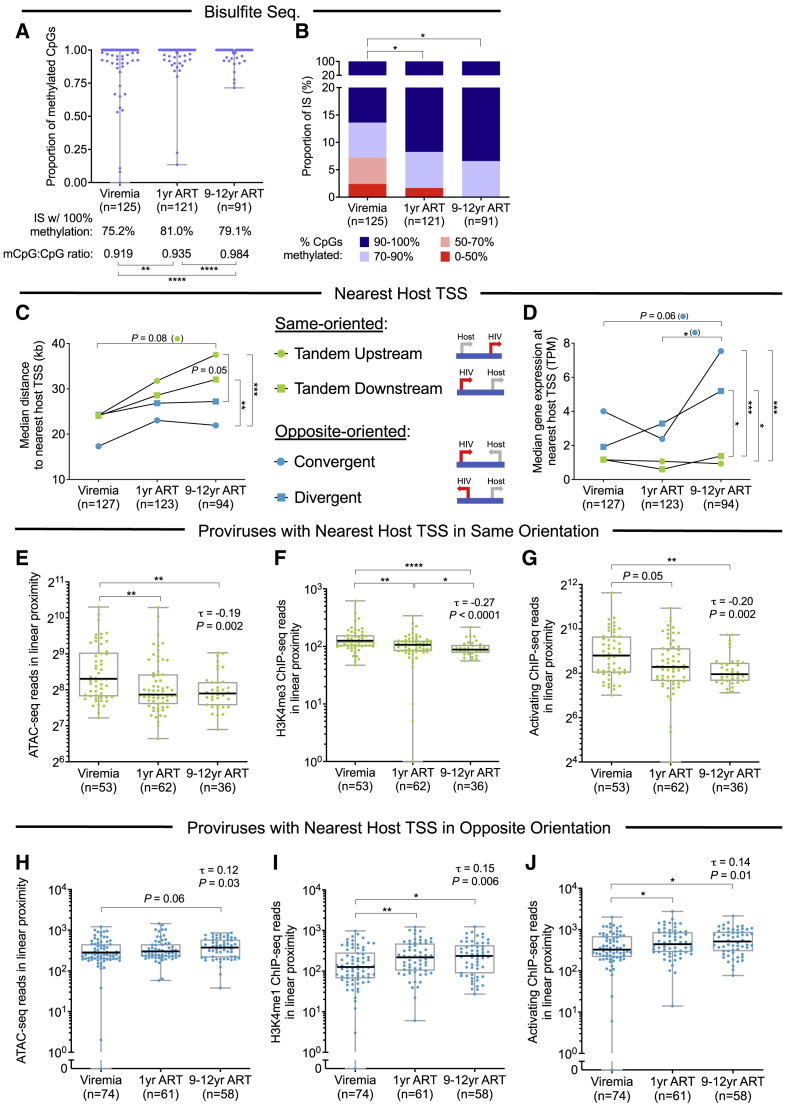
Figure 4.
Longitudinal evolution of proviral integration site features
(A and B) Proportion of methylated CpG (mCpG) residues within 2,500 bp upstream of the HIV-1 5′-LTR promoter for IS. Proportions of IS with 100% upstream CpG methylation and the average ratio of methylated CpGs to total CpGs are also indicated. Proviruses with 0 CpGs within 2,500 bp upstream of the integration site were excluded.
(C) Median distance between proviral IS and the most proximal host transcriptional start site (TSS) with indicated orientation to the proviral sequence.
(D) Median RNA-seq-derived gene expression intensity at nearest host TSS with indicated directional orientation to proviral sequence.
(E–G) Among proviruses in the same directional orientation as the nearest host TSS, plots indicate the longitudinal evolution of ATAC-seq reads (E) and H3K4me3-specific (F) and all activating (H3K4me1, H3K4me3, and H3K27ac) ChIP-seq reads (G) surrounding (±10 kb) proviral IS.
(H–J) Among proviruses in opposite orientation to the nearest host TSS, plots indicate the longitudinal evolution of ATAC-seq reads (H), H3K4me1-specific (I), and all activating (H3K4me1, H3K4me3, and H3K27ac) ChIP-seq reads (J) surrounding (±10 kb) proviral IS.
(E–J) Kendall’s rank correlation coefficients (τ) and corresponding p values are indicated in the upper right of each plot.
(A–J) Longitudinal data from all proviruses in genic regions from study subjects 1, 2, and 5 are included; IS located in chromosomal regions in the ENCODE blacklist (Amemiya et al., 2019) were excluded; clonal IS are counted only once and assigned to the time point contributing the majority of clonal members or to the earliest time point in the case of a tie. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Mann-Whitney U tests, Fisher’s exact tests, or G tests were used for all comparisons).