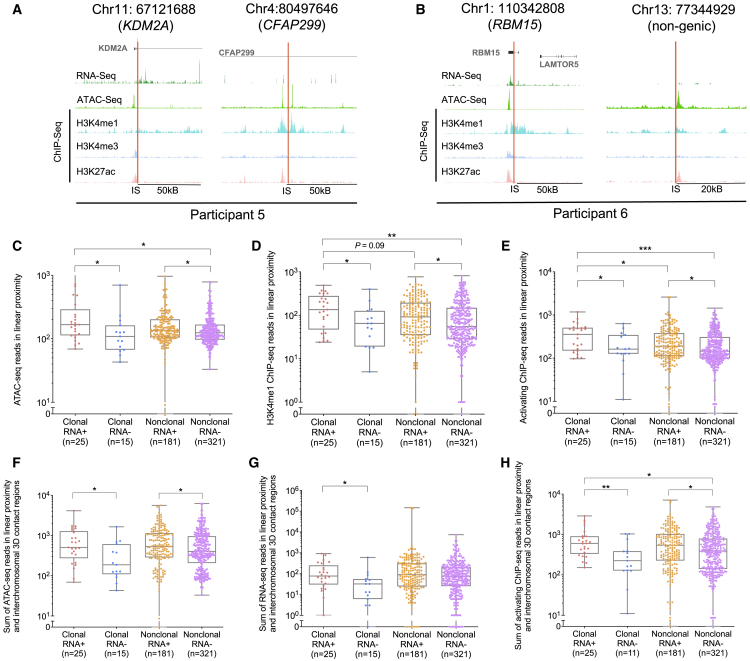
Figure 6.
Epigenetic features of transcriptionally active clonal HIV-1 proviruses
(A and B) Genome browser snapshots reflecting the local chromatin environment surrounding the proviral IS of selected transcriptionally active clonal proviruses from study persons 5 (A) and 6 (B).
(C–E) ATAC-seq (C), H3K4me1-specific ChIP-seq (D), and all activating (H3K4me1, H3K4me3, and H3K27ac) ChIP-seq (E) reads surrounding (±5 kb) the proviral IS of clonal proviruses and of proviruses detected once (here termed “nonclonal”).
(F–H) Sum of ATAC-seq (F), RNA-seq (G), and all activating ChIP-seq (H) reads in linear proximity and 3D interchromosomal contact regions of clonal proviruses and proviruses detected once (“nonclonal”) using Hi-C data at 10 kb binning resolution.
(C–H) HIV-1 Long LTR RNA-expressing proviruses were considered “RNA+”; HIV-1 Long-LTR RNA-negative proviruses are considered "RNA-". Clonal sequences were counted only once; clones were counted as transcriptionally active when at least one member of a clonal cluster had detectable expression of HIV-1 long LTR RNA. IS located in chromosomal regions in the ENCODE blacklist (Amemiya et al., 2019) were excluded. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Mann-Whitney U tests were used for all comparisons).