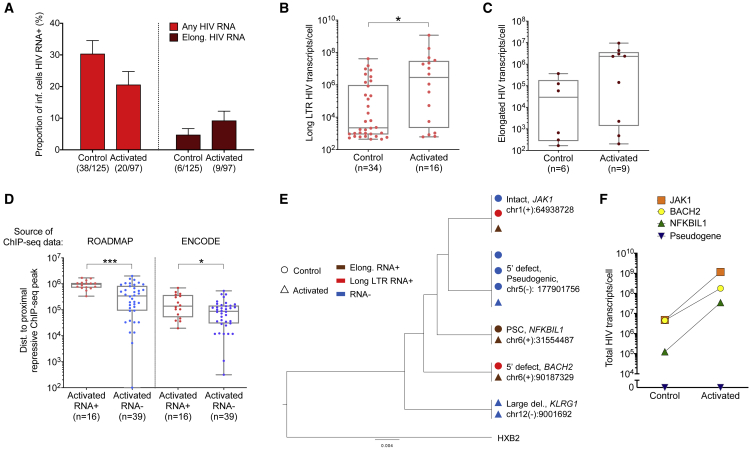
Figure 7.
Transcriptional activity of individual proviruses after in vitro stimulation
(A) Proportion of proviruses producing any HIV-1 RNA or elongated HIV-1 RNA after 12 h of stimulation with PMA/ionomycin or control media.
(B and C) Per-cell levels of HIV-1 long LTR (B) and elongated (C) transcripts from single HIV-1-infected cells after 12 h of stimulation with PMA/ionomycin or control media. Only proviruses with detectable HIV-1 RNA are included.
(D) Chromosomal distance between proviral IS and nearest ChIP-seq peaks corresponding to repressive histone marks (H3K27me3 and H3K9me3) among viral RNA-positive or -negative proviruses stimulated with PMA/ionomycin. IS are annotated with ChIP-seq data from the ROADMAP project (resting primary CD4+ T cells, Kundaje et al., 2015) or from ENCODE (activated primary CD4+ T cells ). IS located in chromosomal regions in the ENCODE blacklist (Amemiya et al., 2019) were excluded.
(E) Phylogenetic tree of clonal proviral species that were detected in stimulated and nonstimulated experimental conditions.
(F) Per-cell levels of total HIV-1 transcripts detected in clonal HIV-1-infected cells analyzed in the presence or absence of stimulation with PMA/ionomycin. (∗p < 0.05, ∗∗∗p < 0.001, Mann-Whitney U tests were used for all comparisons. Error bars represent SEP).