Figure S1.
Technical evaluation of PRIP-seq assay, related to Figure 1
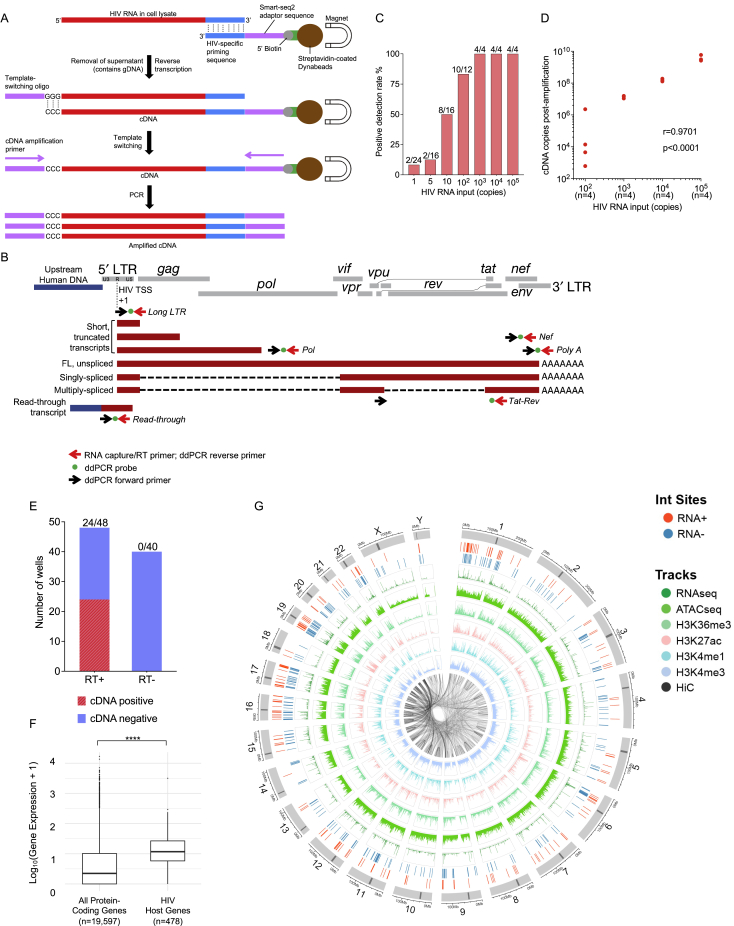
(A and B) Schematic representation of the experimental workflow for isolation, reverse transcription, and amplification of HIV-1 RNA/cDNA (A) and of the primer/probe binding sites for ddPCR-based detection of indicated HIV-1 cDNA products (B).
(C and D) Known HIV-1 RNA copy numbers were serially diluted in 96-well plates and added to cell lysates of 10,000 PBMC from an HIV-1-uninfected person; afterward, a standard PRIP-seq assay was performed. (C) Proportion of wells with detectable HIV-1 cDNA at the indicated number of input HIV-1 RNA copies. (D) Correlation between input HIV-1 RNA copy numbers and numbers of postamplification HIV-1 cDNA copies detectable by the PRIP-seq assay; Spearman correlation coefficient is shown.
(E) Evaluation of possible HIV-1 cDNA contamination by genomic HIV-1 DNA. PRIP-seq was applied to 48 wells, each containing 12,000 PBMC/well from an HIV-infected participant; 40 separate control wells were subjected to the same protocol, except for exclusion of reverse transcriptase from the workflow. Graph demonstrates number of wells with detectable HIV-1 cDNA in samples and controls.
(F) Gene expression intensity (determined by RNA-seq) of all human protein-coding genes compared with host genes harboring proviral IS recovered by PRIP-seq in all study subjects. (∗∗∗∗ p < 0.0001, Mann-Whitney U test).
(G) Circos plot indicating positioning of long LTR RNA-expressing proviruses (RNA+) and transcriptionally silent (RNA-) proviruses relative to genome-wide assessments of indicated transcriptional (RNA-seq), epigenetic (ATAC-seq and ChIP-seq) and three-dimensional chromatin contact (Hi-C) features. Data from all analyzed proviruses for which IS were available are shown.