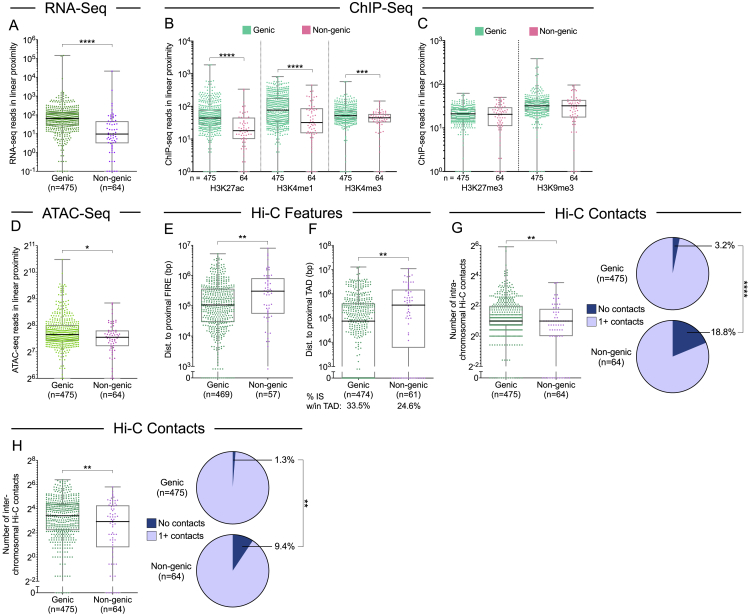
Figure S3.
Chromatin features of HIV-1 proviruses integrated in non-genic DNA, related to Figures 1 and 2
(A–D) Sum of local RNA-seq reads (A), ChIP-seq reads corresponding to activating (B), inhibitory (C) histone modifications, and ATAC-seq reads (D) within 5 kb upstream or downstream of proviral IS in genic versus nongenic locations.
(E and F) Chromosomal distances of proviruses in genic versus nongenic positions to frequently interacting regions (FIREs) (E) and to topologically associated domains (TADs) (F), determined at 10 kb binning resolution of Hi-C data.
(G and H) Numbers of intrachromosomal (G) and interchromosomal (H) contact regions, determined by FiTHiC2-seq (Kaul et al., 2020) (p < 0.05, binning resolution of 20 kb), for proviruses in genic versus nongenic locations. Pie charts reflect proportions of proviruses with no detectable intra- or interchromosomal contacts. (A–H) Clones of proviruses are counted as single datapoints; IS located in chromosomal regions in the ENCODE blacklist (Amemiya et al., 2019) were excluded because of the reduced ability to map next-generation sequencing reads onto repetitive genomic DNA regions. (E and F) Proviral sequences without FIRE annotation by FIREcaller (Crowley et al., 2021) or without TAD annotation by Homer (version 4.10.3) were excluded from the respective analyses. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; Mann-Whitney U tests or Fisher’s exact tests were used for all comparisons).