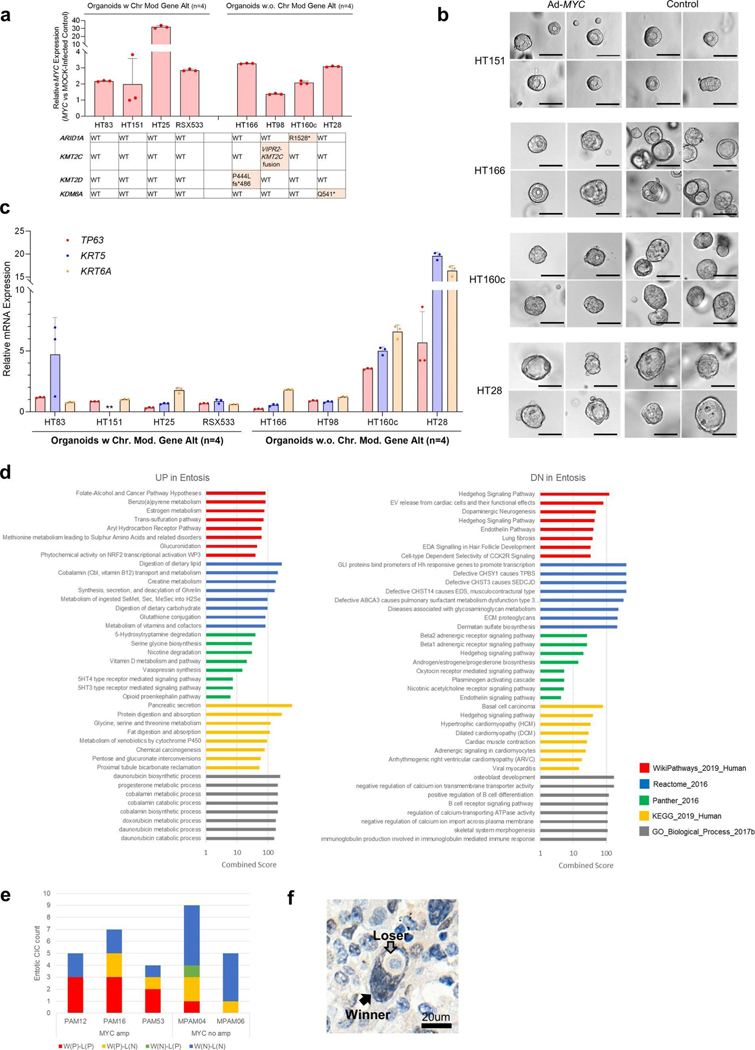
Extended Data Fig. 10 |. Molecular Characteristics in Squamous Feature and Entosis.
(a)-(d) Impact of MYC-overexpression Using PDAC Organoid Models, (a) Overexpression of MYC and alteration status of chromatin modifier genes in eight PDAC organoids. Center value and bar: mean and SD. Three data points in each organoid means technical triplicates of qPCR data, (b) Representative images of PDAC organoids (out of 8 organoids, 64 images). Images of organoids were acquired 5 days post-sorting (10 days total post-infection). No obvious morphological changes were identified between the MYC-infected vs the mock-infected organoids. Scale bar: 50um. (c) Relative mRNA expression of squamous markers (TP63, KRT5and KRT6A) after MYC overexpression. Two PDAC organoids with chromatin modifier mutations (HT160c and HT28) shows higher expression of all three markers whereas no effects are seen in the absence of mutations in these genes. Center value and bar: mean and SD. Three data points in each organoid means technical triplicates of qPCR data. **Appropriate KRT5 in HT151 signal was not detected with qPCR due to low expression, (d) Metabolic pathways are enriched in Entotic cases based on five different databases, (e) MYC expression in Entotic CIC. Winner (MYC positive)-Loser (MYC negative) pattern was identified both MYC amplified and non-amplified cases. W(P)-L(P), W(P)-L(N), W(N)-L(P) and W(N)-L(N) are Winner (MYC positive)-Loser (MYC positive), Winner (MYC positive)-Loser (MYC negative), Winner (MYC negative)-Loser (MYC positive) and Winner (MYC negative)-Loser (MYC negative) patterns respectively, (f) Representative image of Winner (positive)-Loser (negative) pattern (out of 9, 6,1 and 14 images in W(P)-L(P), W(P)-L(N), W(N)-L(P) and W(N)-L(N) patterns).