Abstract
Background
During the current coronavirus disease 2019 (COVID-19) pandemic, many countries require travellers to undergo a reverse transcription-polymerase chain reaction (RT-PCR) testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) before travelling across borders. However, in persons having recovered from COVID-19, RT-PCR positivity can persist for an extended period.
Materials and methods
We describe three cases who sought fit-to-fly certificates in Thailand during the period free of local transmission but were tested positive for RT-PCR for SARS-CoV-2. All had returned from a country with an active outbreak of COVID-19. Their clinical courses are described; positive nasopharyngeal swab samples were processed for viral isolation and whole-genome sequencing (WGS); and serology as well as neutralizing antibody were assessed. The contact tracing was carried out for determining evidence of indigenous transmission among close contacts of those three cases.
Results
All three cases were completely asymptomatic. Chest computerized tomography was not compatible with COVID-19 pneumonia; cell cultures failed to rescue replication-competent virus; WGS revealed fragmented viral genetic material from nasopharyngeal swab samples; and serological tests demonstrated stable levels of antibodies, together with the presence of neutralizing antibody, suggesting past infection with negligible transmission risk. Contact tracing identified no transmission in high-risk close contact individuals.
Conclusion
RT-PCR positivity for SARS-CoV-2 might detect fragmented viral genome. Issuance of a travel certificate in these circumstances is problematic. Serology tests can help to define past infection. A practical acceptable set of guidelines for issuance of a COVID-19 safety travel certification is a necessity.
Keywords: COVID-19, RT-PCR, SARS-CoV-2, Serology, Travel certificate
1. Introduction
An outbreak of coronavirus disease 2019 (COVID-19) emerged in Wuhan, China, in December 2019 and eventually becomes a pandemic [1]. Thailand is the first country outside China reporting a COVID-19 case in early January 2020 [2,3]. Following an initial phase of the first wave of COVID-19 outbreak, local transmission was successfully contained on May 26, 2020 through a country-wide implementation of measures to curb the spread of the disease, namely, lockdown, active case-finding and prompt case isolation [4]. The country later faced the second wave of the local outbreak in mid-December 2020.
An important control measure after the containment of in-country outbreak at the time was a mandatory 14-day quarantine of inbound travellers at the country's ports of entry, during which COVID-19 symptoms are monitored and nasopharyngeal (NP) swabs are collected for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reverse transcription-polymerase chain reaction (RT-PCR) assay between day 3 and 5, with a repeat test one week later [5]. This measure has helped to prevent inbound travellers with previously undiagnosed COVID-19 to introduce the virus into the country.
Many countries currently require a negative RT-PCR test for SARS-CoV-2 for issuance of a ‘fit-to-fly’ health certificate prior to travelling [[6], [7], [8]]. However, in August 2020, when Thailand had no known indigenous COVID-19 transmission, three cases of returning individuals tested positive when seeking a medical certificate for exit travel despite negative RT-PCR quarantine results a few months earlier. The subjects were admitted for clinical and laboratory characterization, and their management is discussed.
2. Materials and methods
2.1. Case definition
The cases were individuals who returned to Thailand and tested negative for SARS-CoV-2 by RT-PCR using an Allplex™ 2019-nCoV Assay (Seegene, Seoul, South Korea) on two occasions during the mandatory 14-day quarantine program, but were tested positive for SARS-CoV-2 by RT-PCR at Ramathibodi Hospital, Bangkok when they sought a fit-to-fly health certificate.
The study protocol was reviewed and approved by the Ethical Clearance Committee on Human Rights Related to Research Involving Human Subjects, Faculty of Medicine Ramathibodi Hospital, Mahidol University (approval no. MURA2020/1533).
2.2. SARS-CoV-2 RT-quantitative (q)PCR assay
RNA was extracted from 200 μL of viral transport media from NP swab samples using MagDEA® Dx kit (Precision System Science, Chiba, Japan) and SARS-CoV-2 RT-qPCR was performed by amplification of SARS-CoV-2 ORF1ab and N fragments using a SARS-CoV-2 Nucleic Acid Diagnostic Kit (Sansure, Changsha, China) [9], with a cycle threshold (Ct) ≤38 considered positive.
2.3. SARS-CoV-2 genome sequencing and analysis
Extracted RNA samples from the RT-qPCR assay were stored in cold chain and processed for whole-genome sequencing (WGS) using an ARTIC-based multiplex enrichment method, as described in Supplementary Methods [10,11]. In short, RNA was reverse transcribed using a SuperScript IV Reverse Transcriptase reaction kit (Thermo Fisher Scientific, Waltham, MA, USA) together with a random hexamer (Thermo Fisher Scientific). Multiplex PCR amplification employing an ARTIC primer set version 3 (Quick 2020) and Platinum SuperFi DNA polymerase (Thermo Fisher Scientific) was performed at 98 °C for 30 s, followed by 35 cycles of 98 °C for 15 s and 65 °C for 5 min. Amplicons following a clean-up by AMPure XP (Beckman Coulter, Brea, CA, USA) were quantified using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Amplicons were pooled and 100 ng were subjected to library preparation (Nextera Flex kit; Illumina, San Diego, CA, USA), a clean-up (AMPure XP kit; Beckman Coulter) and analysis (Fragment Analyzer System; Agilent, Santa Clara, CA, USA; Qubit dsDNA HS assay kit; Thermo Fisher Scientific). An equimolar amount of the libraries was denatured in 0.05 M NaOH at room temperature for 5 min, diluted to 10 pM and sequenced using a Miseq reagent Nano kit version 2 (500 cycles) (Illumina). Variants were identified by a ncov2019-artic-nf pipeline (https://github.com/connor-lab/ncov2019-artic-nf) utilizing default Illumina parameters (variants called by iVar version 1.11 with minimum frequency threshold 0.75 and minimum depth of 10) [12]. Read fragments were mapped to an MN908947.3 reference genome using bwa 0.7.17. Emerging time and the associating lineage of variants were determined by Nextstrain and Global Evaluation of SARS-CoV-2 Sequences (GESS).
2.4. Virus isolation and microneutralization assay
Vero E6 cells (ATCC® CRL-1586™) were maintained in DMEM medium (Gibco; Thermo Fisher Scientific) supplemented with 10% foetal bovine serum (Gibco), 100 U/mL penicillin and 0.1 mg/mL streptomycin at 37 °C under a humidified atmosphere containing 5% CO2. NP swab contents were added to Vero E6 cells and monitored for cytopathic effect (CPE) daily for seven days. A blind passage was taken for every sample. Cells with CPE were subjected to RNA extraction from both culture supernatant and cell pellet and assayed for virus presence by RT-PCR using primers targeting ORF1ab and N (Sansure). Culture experiments were performed in a certified biosafety level 3 (BSL-3) laboratory.
Microneutralizing antibody assays for SARS-CoV-2 were performed in the BSL-3 laboratory. In brief, serum samples were heated at 56 °C for 30 min and two-fold serially diluted with cell culture media, and each dilution was mixed with 100X median tissue culture infective dose (TCID50) and incubated for 1 h at 37 °C. Each mixture was then overlaid on 1 × 104 Vero E6 cells in a well of a 96-well plate, incubated at 37 °C for two days, CPE observed under a light microscope (40x magnification), then cells were washed and fixed with ice-cold methanol/acetone. Infected cells were detected by an enzyme-linked immunosorbent assay (ELISA) employing a rabbit anti-SARS-CoV/SARS-CoV-2 nucleocapsid (N) protein monoclonal primary antibody (Sino Biological, Wayne, PA, USA) and a goat HRP-conjugated anti-rabbit secondary polyclonal antibody (Dako, Glostrup, Denmark). Immunoreactive protein was visualized using 3,3′,5,5′-tetramethylbenzidine (TMB) substrate; neutralizing endpoint is expressed as reciprocal of the highest dilution of serum sample giving mean signal (A450/620nm) < 50% of negative control serum sample. Each experiment was performed in quadruplicate.
2.5. Antibody detection assay
Levels of IgG and IgA against SARS-CoV-2 spike (S) protein were measured using an ELISA automated system (Euroimmun, Luebeck, Germany), IgM and IgG against virus N and S protein using a MAGLUMI 2000 automated chemiluminescence immunoassay (CLIA) (Snibe, Shenzhen, China), IgG against N protein using an Abbott Architect automated CLIA (Lake Forest, IL, USA), and total antibodies against S1 domain of S protein using a VITROS System automated CLIA (Ortho Clinical Diagnostics, Raritan, NJ, USA). Employing Snibe internal reference calibrators, results of IgM and IgG immunoassays are expressed as AU/mL with a positive cut-off ≥1.0. Other immunoassay results are expressed as sample signal/calibrator cut-off (s/co) with positive cut-off ≥1.4 for Abbott anti-N protein IgG assay, positive cut-off ≥1.0 for Ortho anti-S1 protein total antibody assay, and positive cut-off ≥1.1, borderline 0.8–1.1 and negative cut-off ≤0.8 for Euroimmun anti-S protein IgA and IgG assays.
2.6. Contact tracing
The contact tracing was carried out by responsible health officers of the Ministry of Public Health. Every laboratory-confirmed case of COVID-19 case triggers a prompt investigation of their close contacts per protocol [13]. The contacts were interviewed, and the NP swab samples were collected for SARS-CoV-2 RT-PCR test. In addition, sera of some high-risk close contacts were tested for SARS-CoV-2 IgM and IgG (Abbott Architect; Lake Forest, IL, USA). Epidemiological information together with clinical and laboratory data was obtained from the contacts, mainly household members, to determine evidence of indigenous transmission.
3. Results
3.1. Clinical presentations and laboratory investigations
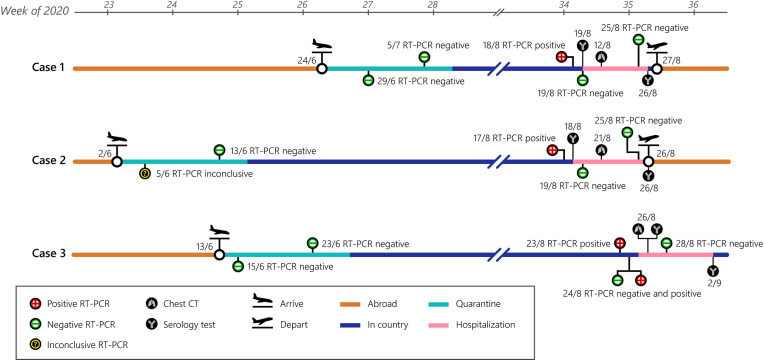
Three Thai females, who worked in the United Arab Emirates (UAE) and travelled back to Thailand in June 2020 without any known COVID-19 symptoms, were quarantined in different Thai-government-assigned facilities for 14 days and tested negative for SARS-CoV-2 by RT-qPCR assay of NP swabs on two occasions prior to returning to their respective home towns. In August 2020, the subjects independently sought a fit-to-fly medical certificate to return to work abroad. SARS-CoV-2 RT-qPCR assays of NP swabs were found to be positive, although all subjects were completely asymptomatic during the intervening period when there was no known local transmission in the country. The subjects were admitted for clinical monitoring and laboratory investigations (Fig. 1 ).
Fig. 1.
Timeline of clinical and laboratory investigations of Cases 1–3. Diagrams depicted are based on interviews, medical records and observations. Dates (day/month) of tests are in 2020.
Case 1 is a 35-year-old healthy woman who remained in lockdown from February to June 2020 owing to the COVID-19 pandemic in UAE and recalled no symptoms of respiratory tract infection or loss of taste or smell during the whole period. Upon returning to Thailand on June 24, 2020, Case 1 was placed under a 14-day quarantine. SARS-CoV-2 RT-PCR tests of NP swabs on June 29 and July 5, 2020 were negative and she was allowed to return to her home town where she remained with her family who were all asymptomatic. When Case 1 sought a fit-to-fly medical certificate, a positive SARS-CoV-2 RT-qPCR test was obtained on August 18, 2020 (Ct values of 38 and 34 for ORF1ab and N, respectively). On admission, Case 1 reported no symptoms; vital signs were within normal ranges; peripheral oxygen saturation was 98–99% while breathing ambient air; chest computerized tomography (CT) showed no lung abnormality; and complete blood count (CBC), creatinine and lactate dehydrogenase (LDH) levels, and liver function tests (LFTs) were within normal ranges.
Case 2 is a 34-year-old female without any co-morbid condition and had discontinued her work in UAE and remained a housewife from February to June 2020 during which she reported no respiratory symptoms, or loss of taste or smell. Case 2 arrived in Thailand on June 2, 2020 and was placed under a 14-day quarantine, remained asymptomatic and SARS-CoV-2 RT-qPCR NP swab test on June 5, 2020 showed weak positive results (Ct values of 37 and 40 for RdRP and N, respectively). However, the second confirmation test was negative, and RT-qPCR test on June 13, 2020 was also negative. Case 2 returned to her home town and stayed with her family who were asymptomatic. On August 17, 2020, SARS-CoV-2 RT-qPCR test of NP swab showed weak positive results (Ct values of 40 for both ORF1ab and N). RT-qPCR test of NP swab performed on the following day showed Ct values of 36 and 32 for ORF1ab and N, respectively. On admission, Case 2 reported no symptoms; vital signs were normal; peripheral oxygen saturation was 98–99% while breathing ambient air; chest CT showed several small non-specific pulmonary nodules in right middle, right lower, left upper, and left lower lobes as well as a few peri-fissural nodules at right major and minor fissures, which were deemed by attending radiologists as non-specific and not typical manifestations of COVID-19 pneumonia; and CBC, creatinine and LDH levels and LFTs were within normal ranges.
Case 3 is a 40-year-old healthy female whose work in UAE was terminated in the middle of March 2020 due to COVID-19 pandemic and remained completely asymptomatic during the time. Case 3 returned to Thailand on June 13, 2020, placed under a 14-day quarantine and SARS-CoV-2 RT-qPCR tests of NP swabs on June 15 and 23, 2020 were negative, and was permitted to return to her home town. On August 23, 2020, SARS-CoV-2 RT-qPCR test of NP swab showed Ct values of 37 and 35 for ORF1ab and N, respectively, and two separate tests conducted on the following day produced, in one test, a negative result for ORF1ab and a Ct value of 38 for N, and, in the other test, Ct values of 38 and 34 for RdRp and N, respectively. On admission, Case 3 reported no symptoms, vital signs were normal; peripheral oxygen saturation was 99–100% while breathing ambient air; chest CT showed a few linear opacities at the lateral segment of right middle lobe and inferior lingular segment, compatible with subsegmental atelectasis or fibrosis; and CBC, creatinine and LDH levels, and LFTs were within normal ranges.
3.2. Serum antibody assays
In the three cases, at hospital admission, anti-SARS-CoV-2 IgG but not IgM was detected, while IgA titres were borderline in Case 1 and positive in Cases 2 and 3 (Table 1 ). Furthermore, all cases possessed neutralizing antibody against SARS-CoV-2. Evaluation of the second serum samples of the cases drawn 7–9 days later showed comparable titres of IgM, IgG and IgA, as well as neutralizing antibodies (Table 1). It is noteworthy that COVID-19 vaccine was not available at the time. Thus, serological test results indicate that the cases were not in an acute phase of infection during the time of hospital admission.
Table 1.
Serology and neutralizing antibody titres of travellers with SARS-CoV-2 RT-PCR positivity during stay in Thailand.
| Case | Date (day/month) | Anti-N/S protein IgM |
Anti-N/S protein IgG |
Anti-N protein IgG Abbott s/co (≥1.4)a | Anti-S1 protein total antibody |
Anti-S protein IgG Euroimmun s/co (≥1.1)a | Anti-S protein IgA |
Microneutralization antibody titre (≥10)a |
|---|---|---|---|---|---|---|---|---|
| Snibe AU/mL (≥1.0)a | Snibe AU/mL (≥1.0)a | Ortho s/co (≥1.0)a | Euroimmun s/co (≥1.1)a | |||||
| # 1 | 19/8 | 0.54 | 1.03 | 3.58 | 27.0 | 1.37 | 0.90 | 320 |
| 26/8 | 0.54 | 1.00 | 3.39 | 31.1 | 1.27 | 0.95 | 160 | |
| # 2 | 18/8 | 0.50 | 2.96 | 2.72 | 98.0 | 2.39 | 1.54 | 80 |
| 26/8 | 0.35 | 2.32 | 2.01 | 106 | 2.03 | 1.62 | 20 | |
| # 3 | 26/8 | 0.33 | 6.44 | 1.57 | 129 | 2.02 | 2.48 | 80 |
| 2/9 | 0.37 | 6.81 | 1.94 | 181 | 1.97 | 2.64 | 80 |
Positive cut-off value. S/co, signal/cut-off ratio.
3.3. Virus rescue assay
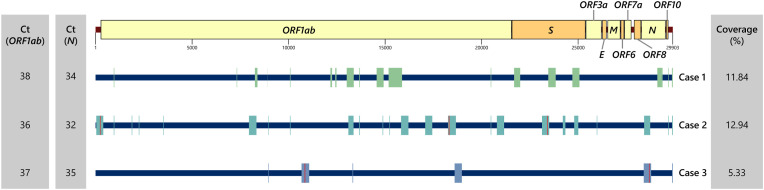
In order to determine if these three cases were able to transmit SARS-CoV-2 to others, viral cultures from NP swabs in Vero E6 cells were performed but no replication-competent virus was obtained. In addition, WGS of NP swab specimens using an ARTIC multiplex enrichment method demonstrated mostly fragmented genomes, with the presence of <15% of the intact genome (Fig. 2 ), suggesting viral RNA detected in NP swabs from the three cases did not come from the intact virus.
Fig. 2.
SARS-CoV-2 genome fragments identified in Cases 1–3. Location of genomic fragment (colored box) from ≥10 amplicon is shown. Vertical red line indicates location of mutation. Ct, cycle threshold. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Virus nucleotide sequencing
Due to modest amounts of sequences obtained from WGS, it was not possible to assign SARS-CoV-2 samples into lineages [14]. Nevertheless, several mutations could be identified. In Case 2, mutations C241U, C18312A, and A23403G were detected. The first and the third mutations were first identified in January 2020 in B.1, a viral lineage found in many parts of the world, including the UAE and Thailand [15]. In Case 3, G10848 and U28688C were found. The latter mutation was detected in B.4, a lineage associated with Middle Eastern countries, including the UAE. Nevertheless, these partial reads should not provide a definite lineage assignment. In addition, there is a possibility that these lineages might be in circulation in Thailand without being detected by the genomic surveillance program.
3.5. Contact tracing
Following hospital admission, household members and other close contacts of the cases were interviewed: 27, 2 and 4 contacts of Cases 1, 2 and 3, respectively. All high-risk close contacts were asymptomatic and SARS-CoV-2 RT-qPCR of NP swabs yielded negative results. Detection of serum IgM and IgG of 21 high-risk close contacts revealed an absence of antibodies reactive to SARS-CoV-2. The remaining high-risk contacts did not consent for blood sample collection. These findings suggest the three cases acquired infection prior to returning to the country and did not come from any local transmission.
3.6. Hospital course and clinical management
All three cases were asymptomatic during their hospital stay and were discharged after 8–9 days. Based on clinical and laboratory data, which were compatible with SARS-CoV-2 infection prior to entry into the country, issuance of a fit-to-fly health certificate was deemed appropriate. Cases 1 and 2 travelled to UAE on August 27 and 26, 2020 respectively, while Case 3 has remained in the country at the time of manuscript preparation.
4. Discussion
Despite being asymptomatic and absence of intact SARS-CoV-2 genome in NP swabs, three Thai individuals returning from a COVID-19 endemic country presented SARS-CoV-2 RT-qPCR positivity in NP swabs some 50 days after clearing a 14-day quarantine period. Hospital examination and laboratory tests provided serological evidence of past infection but not of any current COVID-19 symptoms or with the transmissible virus.
In individuals infected with SARS-CoV-2 virus, specific antibodies develop within 1–2 weeks after the onset of symptoms [[16], [17], [18], [19]]. IgG, IgM and IgA titres rise sharply and reach their plateaus around 2–4 weeks following symptoms onset and then slowly decline, with IgM titre being lower than the other two antibody types. Similarly, neutralizing antibody appears after 7–10 days of symptoms onset and reaches a peak at about one month [19,20]. IgG titre persists longer than IgM and IgA [[17], [18], [19]]. Viral RNA persists for a variable period and has been reported up to 92 days after onset of symptoms [21]. However, the virus can only be isolated from respiratory specimens in mild cases of COVID-19 during the first week of symptoms despite subsequent viral nucleic acid detection in respiratory tract specimens [22]. In addition, the reappearance of viral RNA from swabs obtained from the respiratory tract in recovered COVID-19 patients has been described [[23], [24], [25]]. The underlying mechanism(s) of these findings remain(s) elusive.
Korea Disease Control and Prevention Agency reported RT-PCR positivity in 285 previously confirmed COVID-19 cases discharged from quarantine on days 8–82 after initial symptom onset [23]. No secondary infections were present in 790 contacts. Viral cultures from specimens of 108 re-positive cases were negative, and RT-qPCR Ct values were above 30 in approximately 90% of these samples. Neutralizing antibody was found from the sera obtained on the first serum of all the cases. A second study identified COVID-19 RT-PCR positivity in approximately 14% of discharged cases, all with mild or moderate symptoms at initial diagnosis and younger compared to the overall discharged cases [24]. Neutralizing antibody titres were positive in 98% of the cases. No full-length viral genomes were obtained or could live viruses be rescued from culture. RT-qPCR Ct values were above 30 in 10 out of 11 samples with available data. A third study described 20 asymptomatic cases from 182 COVID-19 patients who re-tested positive after hospital discharge, 14 of whom possessing specific anti-SARS-CoV-2 IgG and IgA [25].
To date, while some countries still face active COVID-19 epidemic, others have managed to control local outbreaks, and among the latter, in order to prevent re-introduction of the virus, returning individuals a number of countries require a ‘fit-to-travel’ SARS-CoV-2 medical certificate from all in-coming individuals, and usually a mandatory 14-day quarantine [[5], [6], [7], [8]]. However, as in the present study, more reports are expected of asymptomatic individuals with a recent history of residing in or travelling through a COVID-19 endemic region testing positive in a SARS-CoV-2 RT-PCR test within a relatively short period following arrival and quarantine clearance. One of the key considerations for a fit-to-fly SARS-CoV-2 medical certificate is to ensure individuals do not unnecessarily put others at risk during travel, upon arrival and within the country. Due to the recent emergence of COVID-19 pandemic, regulations for evaluating individuals with the issuance of fit-to-fly travel certificate has yet to be established; nevertheless, the scale of this pandemic makes this kind of travel evaluation inevitable. At present, RT-PCR is generally accepted as a standard COVID-19 test, but positive RT-PCR results, particularly those with the high Ct value range, in individuals previously infected with the virus do not directly indicate infectivity. Further research on hands-on markers associated with viral infectivity is needed. Additionally, it is conceivable that RT-PCR for a screening purpose in asymptomatic persons without any history of significant exposure might generate a considerable risk of false-positive results, as low pretest probability is expected to produce decreased positive predictive value. Therefore, it may be necessary to repeat testing, especially in samples with high Ct values [26].
The strength of the present study lies with comprehensive characterization (clinical picture, radiological examination, RT-PCR test, WGS study, viral culture, and serology) of cases with positive SARS-CoV-2 RT-qPCR tests in a country during a period with no detectable community-acquired COVID-19. The limitations of the study are (i) no serum specimen of the cases in June 2020 was available to further prove that the infection occurred prior to entry, (ii) inability to obtain respiratory tract specimens (“weak positive” specimen) of Case 2 in June 2020 for genome sequence comparison, (iii) viral genome sequences recovered from all cases were not sufficient to unambiguously determine the origin of the viruses, and (iv) there was a remote possibility that asymptomatic transmission occurred in the country and confounded the results.
5. Conclusion
In summary, this study describes RT-qPCR-based detection of SARS-CoV-2 in NP swabs from three asymptomatic Thais some 50 days after clearance of COVID-19 infection following 14 days of quarantine upon arrival from a COVID-19 endemic country. Hospital admission for thorough physical examinations and laboratory tests cleared all three cases of any evidence of current active infection or presence of transmissible virus but provided serological data of previous viral exposure. Virus genome nucleotide sequencing suggested SARS-CoV-2 was probably that circulating in the previous endemic country of residence, consistent with the transmission-free status of Thailand at the time. With COVID-19 pandemic still persisting in many parts of the world, there will be increased demand for a robust ‘fit-to-fly/travel’ certification, and a practical and harmonized set of guidelines are clearly needed. To fulfil this goal, research on biomarkers or laboratory tests indicative of truly non-infective virus in a traveller should be of the highest priority.
Funding
The study was funded by Better Health Programme; National Research Council of Thailand (NRCT); National Science and Technology Development Agency (NSTDA); Ramathibodi Foundation; and Thailand Center of Excellence for Life Sciences (TCELS).
CRediT authorship contribution statement
Sirawat Srichatrapimuk: Investigation, Writing – original draft. Thanat Chookajorn: Conceptualization, Funding acquisition, Investigation, Writing – original draft. Theerarat Kochakarn: Investigation. Suppachok Kirdlarp: Investigation. Ekawat Pasomsub: Investigation. Wasun Chantratita: Funding acquisition. Sopon Iamsirithaworn: Investigation, Writing – original draft. Mongkol Kunakorn: Investigation, Writing – original draft. Arunee Thitithanyanont: Funding acquisition, Investigation, Writing – original draft. Somnuek Sungkanuparph: Supervision. Angsana Phuphuakrat: Conceptualization, Writing – original draft, All authors have read and approved the final version of the manuscript.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We thank the outbreak investigation teams of Department of Disease Control, Provincial Health Offices and Bangkok Metropolitan Administration. We are grateful to Emeritus Professor Prapon Wilairat for the valuable comments.
References
- 1.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur J Clin Invest. 2020;50 doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinjoy S., Tsukayama R., Chuxnum T., Masunglong W., Sidet C., Kleeblumjeak P., et al. Self-assessment of the Thai Department of Disease Control's communication for international response to COVID-19 in the early phase. Int J Infect Dis. 2020;96:205–210. doi: 10.1016/j.ijid.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada P., Buathong R., Phuygun S., Thanadachakul T., Parnmen S., Wongboot W., et al. Early transmission patterns of coronavirus disease 2019 (COVID-19) in travellers from Wuhan to Thailand, January 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.8. 2000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ministry of Public Health, Thailand. Thailand's experience in the COVID-19 response. https://ddc.moph.go.th/viralpneumonia/eng/file/pub_doc/LDoc9.pdf. [Accessed 28 October 2020].
- 5.Department of Disease Control, Ministry of Public Health, Thailand. Guidance for integrated management of state quarantine facilities. https://ddc.moph.go.th/viralpneumonia/eng/file/guidelines/g_guarantine_190620new.pdf. [Accessed 28 October 2020].
- 6.Embassy of the People's Republic of China in the United States of America. Notice on mandatory COVID-19 negative certificates for China-bound passengers before boarding. http://www.china-embassy.org/eng/visas/zyxx/t1812479.htm. [Accessed 28 October 2020].
- 7.Government of Singapore. Updates to border measures for travellers entering Singapore. https://www.gov.sg/article/updates-to-border-measures-for-travellers-entering-singapore. [Accessed 28 October 2020].
- 8.Thiessen T. COVID travel Europe: here's where you must show negative corona test. https://www.forbes.com/sites/tamarathiessen/2020/08/01/covid-travel-europe-must-show-negative-corona-test/#49f6494c51e5. [Accessed 28 October 2020].
- 9.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2021;27 doi: 10.1016/j.cmi.2020.05.001. 285.e1-285.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., et al. Genomic epidemiology of SARS-CoV-2 in Guangdong province, China. Cell. 2020;181:997–1003. doi: 10.1016/j.cell.2020.04.023. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Department of Disease Control, Ministry of Public Health, Thailand. Guidelines for surveillance and investigation of coronavirus disease 2019 (COVID-19). https://ddc.moph.go.th/viralpneumonia/eng/file/guidelines/g_surveillance_150520.pdf. [Accessed 28 October 2020].
- 14.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 15.Shu H., Wang S., Ruan S., Wang Y., Zhang J., Yuan Y., et al. Dynamic changes of antibodies to SARS-CoV-2 in COVID-19 patients at early stage of outbreak. Virol Sin. 2020 doi: 10.1007/s12250-020-00268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J., Tang X., Bai R., Liang C., Zeng L., Lin H., et al. The kinetics of viral load and antibodies to SARS-CoV-2. Clin Microbiol Infect. 2020;26 doi: 10.1016/j.cmi.2020.08.043. 1690.e1-1690.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Long Q.X., Deng H.J., Hu J., Gao Q.Z., Zhang G.J., et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin Infect Dis. 2021;73:e539–e631. doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banchich A, O'Toole Á. SARS-CoV-2 lineages. https://cov-lineages.org/lineages/lineage_B.1.html. [Accessed 1 Nov 2020].
- 21.Wang J., Hang X., Wei B., Li D., Chen F., Liu W., et al. Persistent SARS-CoV-2 RNA positivity in a patient for 92 days after disease onset: a case report. Medicine (Baltimore) 2020;99 doi: 10.1097/MD.0000000000021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Division of Risk assessment and International cooperation, Korea Disease Control and Prevention Agency. Findings from investigation and analysis of re-positive cases. https://www.cdc.go.kr/board/board.es?mid=&bid=0030. [Accessed 28 October 2020].
- 24.Lu J., Peng J., Xiong Q., Liu Z., Lin H., Tan X., et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59:102960. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan B., Liu H.Q., Yang Z.R., Chen Y.X., Liu Z.Y., Zhang K., et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10:11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. Diagnostic testing for SARS-CoV-2. https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2.]. [Accessed 6 December 2021].