Abstract
The gp120 region of the human immunodeficiency virus type 1 (HIV-1) envelope (env) gene exhibits a high level of genetic heterogeneity across the group M subtypes. The heteroduplex mobility assay (HMA) has successfully been used to assign subtype classifications, but C2V5 primers often fail to amplify African strains. We developed an env gp41-based HMA for which the target sequence is amplified with highly conserved gp41 primers, known to efficiently amplify nucleic acids from HIV-1 group M, N, and O viruses. By using gp41 from a new panel of reference strains, the subtype assignments made by our modified HMA were concordant with those obtained by sequencing and phylogenetic analysis of 34 field strains from 10 countries representing subtypes A to G. Testing of field strains from Nigeria further demonstrated the utility of this modified assay. Of 28 samples, all could be amplified with gp41 primers but only 17 (60.7%) could be amplified with the standard C2V5 primers. Therefore, gp41-based HMA can be a useful tool for the rapid monitoring of prevalent subtypes in countries with divergent strains of circulating HIV-1.
More than two-thirds of the world's human immunodeficiency virus (HIV) infections occur among the population of sub-Saharan Africa, although this population represents only 10% of the global population. While antiretroviral drugs can provide some clinical benefit, the cost and availability of these drugs make their widespread use in developing countries unlikely. Prevention of HIV infection through the development of a safe, efficacious, and affordable vaccine remains the most realistic approach to curtailing the AIDS pandemic.
Extensive genetic characterization of HIV-1 strains from diverse geographic regions has shown that they can be divided into three groups, groups M, N, and O. The group M viruses that are represented by nine “pure” subtypes and six circulating forms (CRFs) are primarily responsible for the global pandemic (21; D. L. Robertson, J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. Sharp, S. Wolinsky, and B. Korber, Letter, Science 228:55–57, 2000). This diversity has presented a challenge to vaccine development. Most strategies for the design of candidate vaccines include immunogens specific for the predominant subtypes and the CRFs of the HIV-1 isolates occurring within a country or geographic region.
The heteroduplex mobility assay (HMA) is a rapid and simple laboratory method for the subtyping of HIV-1 isolates (5, 6) and has been widely used to genetically characterize HIV-1 strains worldwide (1, 3, 13, 24). However, because of the broad heterogeneity within the gp120 region of the env gene and the continued evolution of this region over time (2, 11, 14, 16, 25), increasing numbers of global strains are unamplifiable with the env C2V5 set of PCR primers that were developed for the current HMA kit (19, 23). In an attempt to resolve this problem, we modified the existing HMA by using PCR primers specific for a highly conserved region of gp41. These primers allow the amplification of HIV-1 groups M (subtypes A to H), N, and O and simian immunodeficiency virus strain cpz (SIVcpz) with an efficiency of 95% (26, 27). Furthermore, sequence-based phylogenetic analysis of this region of gp41 allows concordant subtype assignment with the C2V3C3 region in nonrecombinant viruses (19). The HMA was then adapted for comparison of the electrophoretic mobilities of a region of gp41 for subtype determinations.
MATERIALS AND METHODS
Field sample collection.
Whole blood was collected in Nigeria from HIV-infected persons throughout Nigeria coming to local hospitals for blood donations, premarital screening, medical checkups, and AIDS-related illnesses and placed directly into Vacutainer CPT tubes (Becton and Dickinson, Franklin Lakes, N.J.). The samples were sent in cold boxes to the Department of Human Virology at the National Institute for Pharmaceutical Research and Development in Abuja, Nigeria, for centrifugation according to the manufacturer's recommendations. Following removal of all patient identifiers, the centrifuged blood tubes were shipped at ambient temperature to the Centers for Disease Control and Prevention, Atlanta, Ga., where they were processed and analyzed by molecular biology-based assays.
DNA extraction and PCR amplification.
HIV infection status was determined by using the Genetic Systems (Redmond, Wash.) rLAV HIV-1 EIA and was confirmed by Western blotting with HIV Blot 2.2 (Genelabs Diagnostics Pte Ltd., Singapore). Proviral DNA from peripheral blood mononuclear cells of the HIV-infected persons was extracted with an automated DNA extractor (Organon Teknika, Durham, N.C.). From 0.5 to 1 μg of DNA from each specimen was subjected to first-round PCR with the ED5 and ED12 primers supplied with the HMA kit (AIDS Reagent Repository, National Institutes of Health, Bethesda, Md.) in a final volume of 100 μl. An aliquot of 5 to 10 μl from the primary PCR was used with the inner primer pair ES7-ES8 for a secondary PCR to obtain an approximately 700-bp PCR product spanning the C2V5 region of gp120. The conditions were as described in the kit. The primary and nested PCR primers for the gp41 region were gp40F1-gp41R1 and gp46F2-gp48R2, respectively, as described previously (19, 26, 27).
HMA.
For C2V5- and gp41-based HMAs, the following conditions were used: 5 μl of the second-round PCR product (approximately 100 to 250 ng of DNA) was mixed with 5 μl of PCR-amplified subtype reference DNA (approximately 100 to 250 ng) or water and 2 μl of TBE (Tris-borate-EDTA) gel buffer (Novex, San Diego, Calif.). For the C2V5-based HMA we used the reference DNA included in the kit, whereas for the gp41-based HMA we prepared subtype reference DNA by amplification of 444 bp of the gp41 region from subtype reference strains (see Table 1). For heteroduplex formation of both C2V5 and gp41, the samples were heated to 94°C for 2 min and chilled by immediately placing the tubes on ice. The C2V5 and gp41 heteroduplexes and homoduplexes were separated by electrophoresis on a 6% polyacrylamide gel (1× TBE at 200 V for 1 h) by using an XCell II Mini-cell (8 cm by 8 cm by 1 mm; Novex) and were visualized by ethidium bromide staining. The strong bands at the bottoms of the gels (Fig. 1) represent homoduplexes. The subtype assignment of an unknown sample is based on evaluation of the mobilities of heteroduplexes formed by the unknown sample with a set of reference subtypes. Heteroduplexes formed between the unknown sample and the most closely related sequences exhibit the fastest mobilities. Therefore, heteroduplexes formed with the set of reference samples (e.g., B1, B2, and B3) from the assigned subtype (e.g., B) should have markedly faster mobilities than those formed with other subtypes, although the degree of this distinction may be subjective. If the subtype assignment cannot be resolved by HMA, the PCR-amplified product may be further characterized through sequencing and phylogenetic analysis (1, 3, 5, 6, 13, 24).
TABLE 1.
Subtype reference strains used for gp41-based HMA
| Codea | HIV-1 strain | Country of origin | Cloned fragment |
|---|---|---|---|
| A1 | RW20b | Rwanda | gp160 |
| A2 | 3675c (CRF02_AG) | Nigeria | gp160 |
| B1 | BR20b | Brazil | gp160 |
| B2 | TH14b (B′) | Thailand | gp160 |
| B3 | SF164b | United States | 6.6 kb of the 3′ end |
| C1 | MW959d | Malawi | gp160 |
| C2 | ZM651d | Zambia | Full genome |
| D1 | ZR001d | DRCe | gp160 |
| D2 | UG021d | Uganda | gp160 |
| E1 | TH22b | Thailand | gp160 |
| E2 | TH06b | Thailand | gp160 |
| F1 | BR020d | Brazil | gp160 |
| F2 | BR029d | Brazil | gp160 |
| G1 | UG975d | Uganda | gp160 |
| G2 | RU131d | Russia | gp160 |
| H | CF056d | CARf | gp160 |
Letters represent the HIV-1 subtype; numbers indicate different reference strains.
AIDS Research and Reference Reagent Program, National Institutes of the Health, Bethesda, Md.
Henry M. Jackson Foundation, Rockville, Md. (Francine E. McCutchan).
University of Alabama, Birmingham (Feng Gao).
DRC, Democratic Republic of Congo (formerly Zaire).
CAR, Central African Republic.
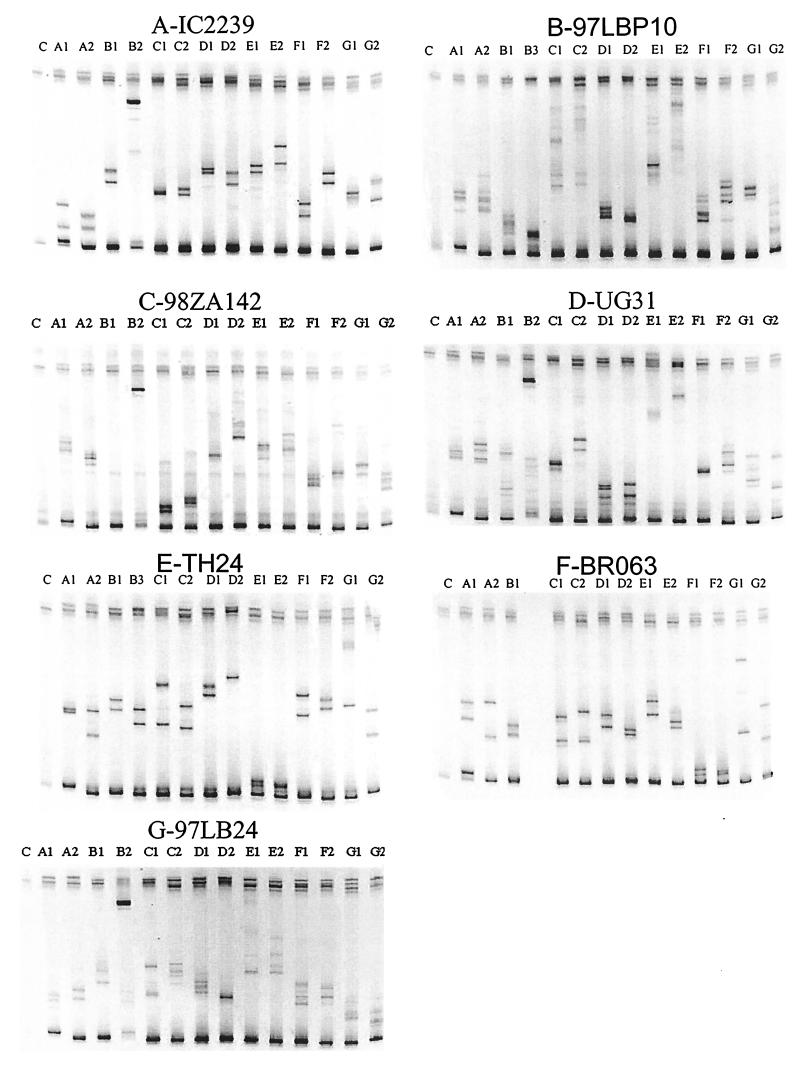
FIG. 1.
HIV-1 subtyping by gp41-based HMA. The subtypes of HIV-1 strains IC2239, UG31, TH135, TH24, 98ZA142, BR063, and 97GLB24 identified are marked with the prefixes A, B, C, D, E, F, and G, respectively. The designations A1, A2, B1, B2, B3, C1, C2, D1, D2, E1, E2, F1, F2, G1 and G2 indicate the subtype reference strains used in the formation of the heteroduplex; C, control.
Sequencing.
Samples were sequenced directly from purified gp41 DNA (QIA-Quick-Spin PCR purification columns; Qiagen Inc., Chatsworth, Calif.) by using the PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems/Perkin-Elmer, Foster City, Calif.) and the halfBD Dye Terminator Reagent (Genpak Inc., Stony Brook, N.Y.) on an automated sequencer (Applied Biosystems Inc., Foster City, Calif.).
Phylogenetic analysis.
The sequences were aligned by use of the CLUSTAL multiple-sequence-alignment program (10). After elimination of positions containing gaps, the aligned sequences of 354 bp were analyzed by the neighbor-joining method with nucleotide distance datum sets calculated by Kimura's two-parameter approach, included in the PHYLIP package (version 3.5c) (8). The stability of the tree topology was tested by pruning, which consists of the removal of one species from the alignment and rerunning of the phylogenetic analysis. The following reference sequences of group M viruses were included in the phylogenetic analysis: subtype A, 92UG037.1 and U455; subtype B, MN, SF2, and 93TH067A (Thai B = B′); subtype C, IN11246, D747, and 92BR025.8; subtype D, 94UG114, NDK, ELI, and 84ZR085.1; subtype E, CM240, 93TH253.3, and 90CF402.1; subtype F, BZ123 and 93BR020.1; subtype G, LBV217; and subtype H, 90CF56.1. The HIV-2 ROD sequence was used as an outgroup.
RESULTS
Purified proviral DNA from 28 HIV-infected persons was subjected to amplification with the primers supplied with the standard C2V5-based HMA kit. DNA from only 17 (60.7%) was successfully amplified. Due to the poor efficiency of amplification of the C2V5 region of env in these Nigerian samples with the PCR primers, we tried amplification with our recently developed gp41 primers, which amplify HIV-1 isolates of groups M, N, and O and SIVcpz (26, 27). The DNA in all 28 (100%) samples was successfully amplified with these primers. Of the 11 samples whose DNA was amplified with the gp41 primers but not the C2V5 primers, 8 (72%) were later identified to contain subtype G viruses. In total, only 5 (38%) of the 13 subtype G strains amplified with gp41 primers could be amplified with the C2V5 primers.
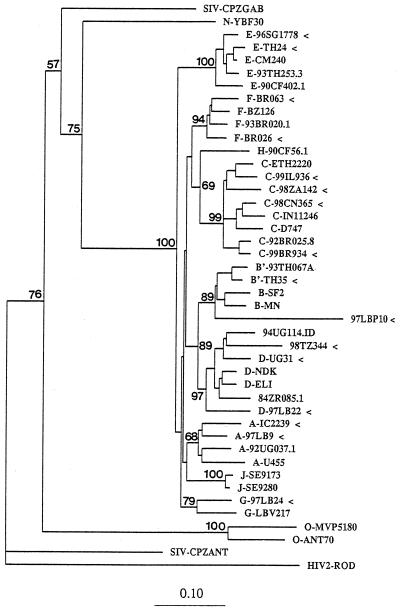
To test whether the gp41 DNA fragment could be used in a standard HMA, we obtained HIV-1 reference clones from subtypes A to H for our gp41-based assay (Table 1). In a proof-of-concept experiment, we used 34 HIV-1 group M field strains representing subtypes A (n = 6), B (n = 4), C (n = 7), D (n = 6), E (n = 5), F (n = 3), and G (n = 3) from 10 countries (Lebanon, Uganda, Tanzania, Thailand, China, South Africa, Israel, Brazil, Ivory Coast, and Singapore) for gp41-based HMA typing. All 34 strains were amplified by PCR with the gp41 primers, and a standard HMA protocol was used to determine the electrophoretic mobilities of the heteroduplexes formed with gp41 PCR-amplified DNA from reference strains of subtypes A to G (Table 1). In addition, the gp41 DNA fragments from our field strains were sequenced and used for phylogenetic analysis. All 34 gp41-based HMA subtype assignments were concordant with the subtype classifications derived from phylogenetic analysis. An example of the gp41-based HMA classification of six field strains of HIV-1 into subtypes A (IC2239, Ivory Coast), B (97LBP10, Lebanon), C (98ZA142, South Africa), D (UG31, Uganda), E (TH24, Thailand), F (BR063, Brazil), and G (97LB24, Lebanon) is presented in Fig. 1; and the phylogenetic assignments of these gp41 sequences are shown in Fig. 2.
FIG. 2.
Phylogenetic classification of env gp41 HIV-1 sequences (arrowheads) from representative group M subtype A to G strains obtained worldwide (GenBank accession numbers AF43895 to AF343910). Sequences denoted with the abbreviations LB, UG, TZ, TH, CN, ZA, IL, BR, IC, and SG were from Lebanon, Uganda, Tanzania, Thailand, China, South Africa, Israel, Brazil, Ivory Coast, and Singapore, respectively. The numbers before those abbreviations indicate the year of specimen collection. Values on the branches represent the percentages of 100 bootstrap replicates. HIV-1 subtypes and groups are marked with the prefixes A, B, C, D, E, F, G, H, and J and the prefixes N and O, respectively. The scale bar indicates the evolutionary distance of 0.10 nucleotides per position in the sequence. Vertical distances are for clarity only.
Finally, viral DNA from 28 HIV-positive Nigerians was amplified with the gp41 primers. All specimens tested by the gp41-based HMA gave unambiguous subtype classifications, and none migrated as fast as recombinant CRF02_AG. The gp41-based sequencing and phylogenetic analyses further confirmed the classifications with 15 subtype A and 13 subtype G viruses detected (data not shown). Overall, all the designations obtained by HMA and by sequencing and phylogenetic analyses were concordant for subtypes A, B, C, D, E, F, and G. While we did not have any strains specifically from North America, we had four subtype B viruses from Lebanon, Thailand, and Brazil. Since subtype B viruses amplify quite well with the conventional C2V5 primers and the gp41 primers were developed for samples that are more difficult to amplify, we believed that use of the four subtype B viruses from other countries was more appropriate. Nonetheless, our phylogenetic analysis confirmed that two of the four subtype B strains, those from Brazil and Lebanon, that we used in our proof-of-concept HMA study were typical of those found in North America. Likewise, subtype A strains from Ivory Coast, Nigeria, and Lebanon, subtype C strains from Israel, South Africa, China, and Brazil, subtype D strains from Tanzania and Uganda, subtype E strains from Thailand and Singapore, subtype F strains from Brazil, and subtype G strains from Nigeria and Lebanon were assigned to the correct subtype by HMA, regardless of their geographic origins.
DISCUSSION
Most current HIV-1 vaccine development strategies use immunogens from locally prevalent subtypes. The need for a rapid, yet simple molecular biology-based screening test for the subtyping of group M viruses and the high level of genetic diversity that characterizes African strains led to the development of the gp41-based HMA. Nigeria is a country with a high number of HIV-1 infections caused by strains of subtypes G and A and multiple AG recombinants, including IbNG strain-like recombinant CRF02_AG (4, 15, 20). From analysis of our Nigerian strains, it appears that PCR amplification of subtype G viruses is not readily achieved with the C2V5 primers included in the original HMA kit. This is most likely due to the greater heterogeneity in the region of env to which the C2V5 primers anneal. This conclusion is supported by the more efficient amplification of the Nigerian strains with PCR primers specific for a highly conserved portion of gp41.
Thus, we modified the standard C2V5-based HMA by replacing the C2V5 PCR primers with highly conserved primers that anneal to the gp41 region of env and by changing the HMA subtype reference strains used. The gp41-based HMA does not require, however, alteration of experimental conditions, studies that used as reported in modifications of other gene regions (7, 9, 18, 22). Moreover, the gp41-based HMA generates easy-to-read electrophoretic profiles after running of the heteroduplexes with subtype reference strains. The success of the gp41-based HMA is likely due to the more conserved nature of this region of env, allowing one to avoid the genetic “noise” caused by the greater levels of nucleotide substitutions and sequence length polymorphisms that characterize the highly variable C2V5 region.
In the present investigation, we have documented that a conserved region in transmembrane protein gp41 in the envelope of HIV-1 can be used for rapid genetic characterization of viral strains into env subtypes A to G, regardless of their geographic origin. Therefore, our HMA may detect all “pure” subtypes and some circulating recombinant forms. The gp41-based assay detects env subtype E viruses, which all belong to CRF01_AE (20, 21). Also, the gp41-based HMA should discriminate between the AG recombinant CRF02_AG (IbNG-like) and “pure” env subtype A, because the PCR-amplified fragment of gp41 includes the mosaic region in which part of the subtype A sequence is replaced with a subtype G sequence (4, 20). This recombinant pattern was first reported in a Nigerian strain (strain IbNg) that was originally identified as a subtype A virus (12). The reference strains used in our gp41-based HMA include both a pure subtype A strain (Table 1, strain A1) and a CRF02_AG recombinant (Table 1, strain A2). It has been reported that CRF02_AG-like strains may cause up to 84% of all infections in West Africa (17). Screening for CRF02_AG-like strains is strongly recommended in future epidemiologic studies to define the role of these viruses in the global pandemic. Ongoing studies in our laboratory indicate that the gp41-based HMA will provide a rapid yet accurate molecular biology-based screening tool for monitoring the spread of CRF02_AG-like infections. The remaining four CRFs (CRF03_AB, CRF04_cpx, CRF05_DF, and CRF06_cpx) (Robertson et al., Letter) could not be detected only by the gp41-based HMA. However, combining the recently reported gag-based HMAs with an env-based HMA may provide a rapid approach to determining a minimum estimate of the number of recombinant viruses circulating within a country (9).
In conclusion, we have found that our gp41-based adaptation of the current env C2V5-based HMA provides a powerful approach for the rapid subtyping of HIV-1 group M viruses worldwide. Furthermore, group M viruses can be classified by HMA into at least seven subtypes, the same number of subtypes that have previously been identified by the C2V5-based HMA. However, the gp41-based HMA allows the subtype classification of a larger number of HIV-1 strains, including CRF02_AG. As our knowledge of the diversity of the HIV strains within a geographic region increases, it will be a simple matter to modify or add additional subtype reference strains for local HIV-1 surveillance studies. Finally, as an understanding of the prevalence of nonrecombinant, as well as recombinant, viruses will be crucial for the development of future HIV vaccines, diagnostics, and treatment strategies, the newly developed gp41-based HMA will facilitate the performance of molecular biology-based surveillance for HIV-1 subtypes in countries preparing for vaccine trials.
REFERENCES
- 1.Bachmann M H, Delwart E L, Shpaer E G, Lingenfelter P, Singal R, Mullins J I. Rapid genetic characterization of HIV type 1 strains from four World Health Organization-sponsored vaccine evaluation sites using a heteroduplex mobility assay. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retrovir. 1994;10:1345–1353. doi: 10.1089/aid.1994.10.1345. [DOI] [PubMed] [Google Scholar]
- 2.Balfe P, Simmonds P, Ludlam C A, Bishop J O, Brown A J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990;64:6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobkov A, Cheingsong-Popov R, Garaev M, Rzhaninova A, Kaleebu P, Beddows S, Bachmann M H, Mullins J I, Louwagie J, Janssens W, van der Groen G, Mc Cutchan F, Weber J. Identification of an env G subtype and heterogeneity of HIV-1 strains in the Russian Federation and Belarus. AIDS. 1994;8:1649–1655. doi: 10.1097/00002030-199412000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 5.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 6.Delwart E L, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl. 1995;4(Suppl. 5):S202–S216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 7.Diaz R S, de Oliveira F, Pardini R, Operskalski E, Mayer A J, Busch M P. HIV type 1 tat gene heteroduplex mobility assay as a tool to establish epidemiologic relationships among HIV type 1-infected individuals. AIDS Res Hum Retrovir. 1999;15:1151–1156. doi: 10.1089/088922299310241. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP: phylogenetic inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Heyndrickx L, Janssens W, Zekeng L, Musonda R, Anagonou S, van der Auwera G, Coppens S, Vereecken K, De Witte K, van Rampelbergh R, Kahindo M, Morison L, McCutchan F E, Carr J K, Albert J, Essex M, Goudsmit J, Asjö B, Salminen M, Buvé A, van der Groen G Study Group on Heterogeneity of HIV Epidemics in African Cities. Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J Virol. 2000;74:363–370. doi: 10.1128/jvi.74.1.363-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 11.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Brown A J. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard T M, Olaylele D O, Rasheed S. Sequence analysis of the glycoprotein 120 coding region of a new HIV type 1 subtype A strain (HIV-1IbNG) from Nigeria. AIDS Res Hum Retrovir. 1994;10:1755–1757. doi: 10.1089/aid.1994.10.1755. [DOI] [PubMed] [Google Scholar]
- 13.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuiken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:5704. [PMC free article] [PubMed] [Google Scholar]
- 15.McCuthan F E, Carr J K, Bajani M, Sunders-Buell E, Hurry T O, Stoeckli T C, Robbins K E, Gashau W, Nasidi A, Janssens W, Kalish M L. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254:226–234. doi: 10.1006/viro.1998.9505. [DOI] [PubMed] [Google Scholar]
- 16.Michael N L, Davis K E, Loomis-Price L D, VanCott T C, Burke D S, Redfield R R, Birx D L. V3 seroreactivity and sequence variation: tracking the emergence of V3 genotypic variation in HIV-1-infected patients. AIDS. 1996;10:121–129. doi: 10.1097/00002030-199602000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Montavon C, Toure-Kane C, Liegeois F, Mpoudi E, Bourgeois A, Vergne L, Perret J-L, Boumah A, Saman E, Mboup S, Delaporte E, Peeters M. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr. 2000;23:363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- 18.Novitsky V, Arnold C, Clevley J P. Heteroduplex mobility assay for subtyping HIV-1: improved methodology and comparison with phylogenetic analysis of sequence data. J Virol Methods. 1996;59:61–72. doi: 10.1016/0166-0934(96)02014-9. [DOI] [PubMed] [Google Scholar]
- 19.Pieniazek D, Young C, Lal R B. Phylogenetic analysis of the gp41 envelope of HIV-1 groups M, N, and O strains provides an alternate region for subtype determination. In: Korber B, Foley B, McCutchan F, Mellors J, Hahn B H, Sodroski J, Kuiken C, editors. Human retroviruses and AIDS 1998. Los Alamos, N.M: Los Alamos National Laboratories; 1998. pp. III112–III118. [Google Scholar]
- 20.Robertson D L, Gao F, Hahn B H, Sharp P M. Intersubtype recombinant HIV-1 sequences. In: Korber B, Foley B, Leitner T, McCutchan F, Hahn B H, Mellors J, Myers G, Kuiken C, editors. Human retroviruses and AIDS 1997. Los Alamos, N.M: Los Alamos National Laboratories; 1997. pp. III25–III30. [Google Scholar]
- 21.Robertson D L, Anderson J P, Bradac J A, Carr J K, Foley B, Funkhouser R K, Gao F, Hahn B H, Kalish M L, Kuiken C, Learn G H, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp P, Wolinsky S, Korber B. HIV-1 nomenclature proposal: a reference guide to HIV-1 classification. In: Kuiken C L, Foley B, Hahn B H, Korber B, McCutchan F, Marx P A, Mellors J W, Mullins J I, Sodroski J, Wolinksy S, editors. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1999. pp. 492–505. [Google Scholar]
- 22.Tatt I D, Barlow K L, Clewley J P. A gag gene heteroduplex mobility assay for subtyping HIV-1. J Virol Methods. 2000;87:41–51. doi: 10.1016/s0166-0934(00)00146-4. [DOI] [PubMed] [Google Scholar]
- 23.Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasi C, Herring B, Raktham S, Vanichseni S, Mastro T D, Young N L, Rubsamen-Waigmann H, von Briesen H, Kalish M L, Luo C C, Pau C P, Baldwin A, Mullins J I, Delwart E L, Esparza J, Heyward W L, Osmanov S. Determination of HIV-1 subtypes in injecting drug users in Bangkok, Thailand, using peptide-binding enzyme immunoassay and heteroduplex mobility assay: evidence of increasing infection with HIV-1 subtype E. AIDS. 1995;9:843–849. doi: 10.1097/00002030-199508000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Wolfs T F, de Jong J J, Van den Berg H, Tijnagel J M, Krone W J, Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci USA. 1990;87:9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Pieniazek D, Oven S M, Fridlund C, Nkengasong J, Mastro T D, Rayfield M A, Downing R, Biryawaho B, Tanuri A, Zekeng L, van der Groen G, Lal R B. Plasma detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers J. Clin Microbiol. 1999;37:2581–2586. doi: 10.1128/jcm.37.8.2581-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Dash B, Simon F, van der Groen G, Pieniazek D, Gao F, Hahn B H, Folks T M, Lal R B. Detection of diverse HIV-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primers. J Infect Dis. 2000;181:1791–1795. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]