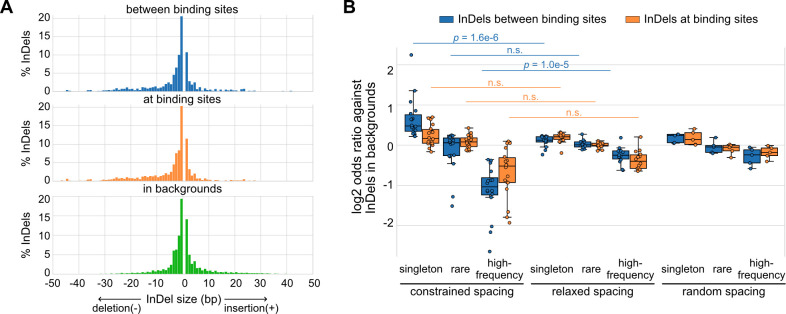
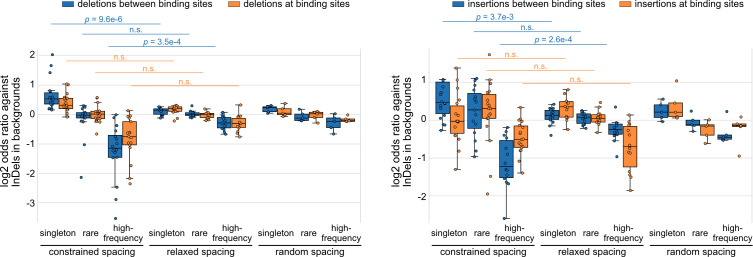
Figure 2. Naturally occurring insertions and deletions (InDels) in human populations.
(A) Size distributions of human InDels within different regions. (B) Log2 odds ratios for different categories of InDels. Each dot represents a transcription factor (TF) pair with corresponding spacing relationship. Mann–Whitney U test was used to compare the odds ratios between different spacing relationships. Non-significant (n.s.) if p-value is larger than 0.01.
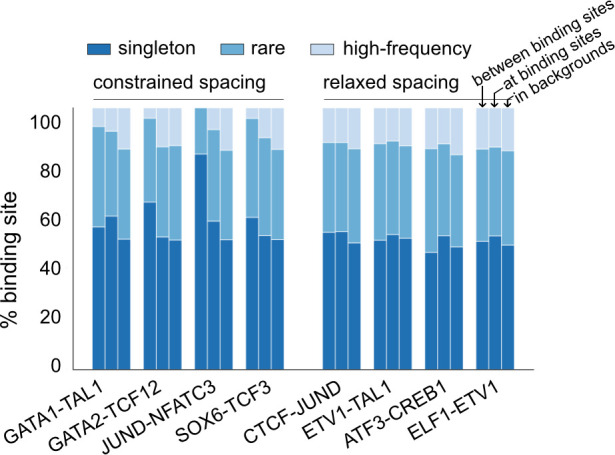
Figure 2—figure supplement 1. Composition of insertions and deletions (InDels) with different allele frequency (AF) for representative transcription factor (TF) pairs.