Figure 6.
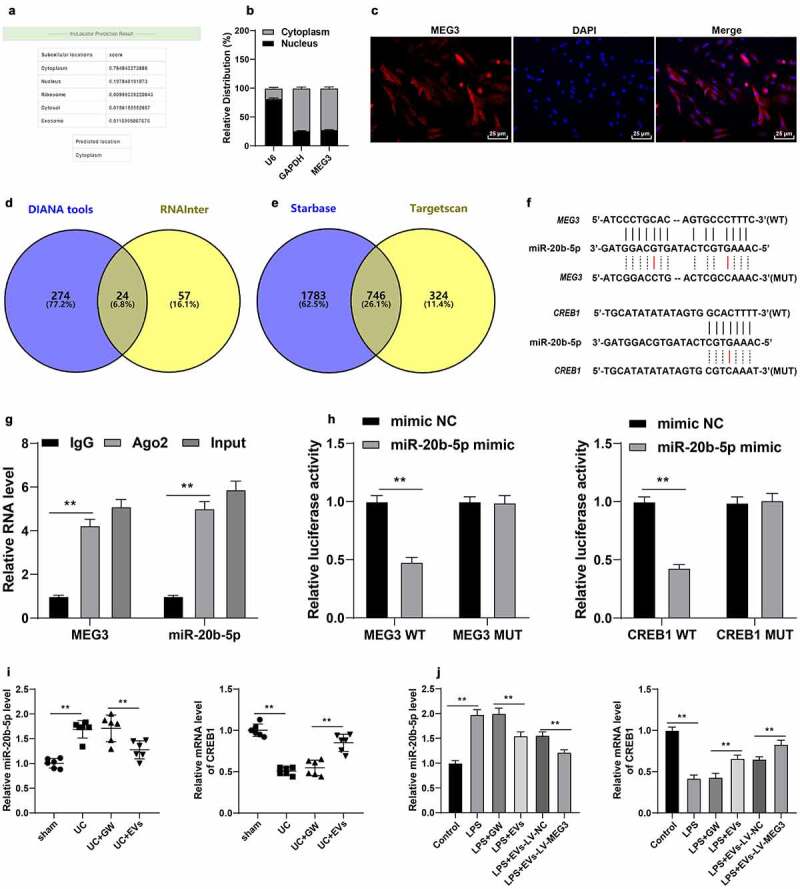
M2-EV-carried MEG3 competitively bound to miR-20b-5p to promote CREB1 transcription. a: The cellular localization of MEG3 was predicted through the LncLocator database (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). b-c: The location of MEG3 in YAMC cells was confirmed using nuclear/cytosol fractionation assay (b) and RNA FISH (c). d: The downstream miRNAs of MEG3 were predicted through the DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) and RNAInter (http://www.rna-society.org/rnainter/). e: The downstream mRNAs of miR-20b-5p were predicted through the Starbase (http://www.targetscan.org/vert_71/) and TargetScan (http://starbase.sysu.edu.cn/index.php). g-h: The binding relationships between miR-20b-5p and MEG3, and miR-20b-5p and CREB1 were analyzed using RIP (g) and dual-luciferase assay (h). i-j: miR-20b-5p and CREB1 expression in cells and tissues were detected using RT-qPCR. N = 6. The experiment was repeated 3 times independently. Data in panels B/G/H/J are presented as mean ± standard deviation. Data in panels I/J were analyzed using one-way ANOVA, and data in panels G/H were analyzed using two-way ANOVA, followed by Tukey’s multiple comparisons test or Sidak’s multiple comparisons test, **p < 0.01
