Figure 4.
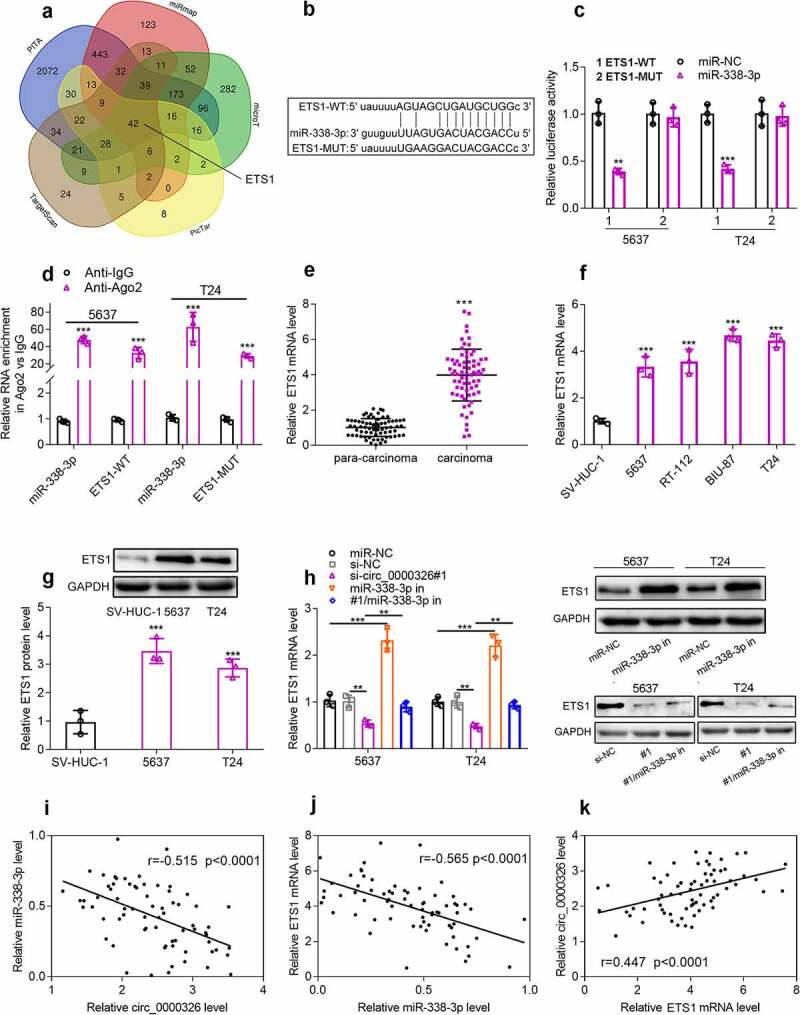
Circ_0000326 could target miR-338-3p to regulate ETS1 expression
(a). Venn diagram of mRNAs containing the potential binding site to miR-338-3p by searching TargetScan, PicTar, PITA miRmap, microT and miRDB databases. (b). StarBase database was employed to predict the binding site between miR-338-3p and ETS1. (c). Luciferase reporter assay confirmed that miR-338-3p could negatively regulate the luciferase activity of circ_0000326-WT, rather than circ_0000326-Mut. (d). RIP assay showed that miR-338-3p and ETS1 were all enriched in Anti-Ago2 group compared with Anti-IgG group. (e-g). qRT-PCR and Western blot showed that ETS1 expression was notably up-regulatedin BC tissues and cells. (h). qRT-PCR and Western blot showed that miR-338-3p inhibitor counteracted the inhibitory effect of circ_0000326 knockdown on ETS1 expression. (i-k). Pearson correlation analysis was used to analyze the relationships between circ_0000326 and miR-338-3p, miR-338-3p and ETS1 mRNA, as well as miR-338-3p and ETS1 mRNA in BC.**P < 0.01 and ***P < 0.001.
