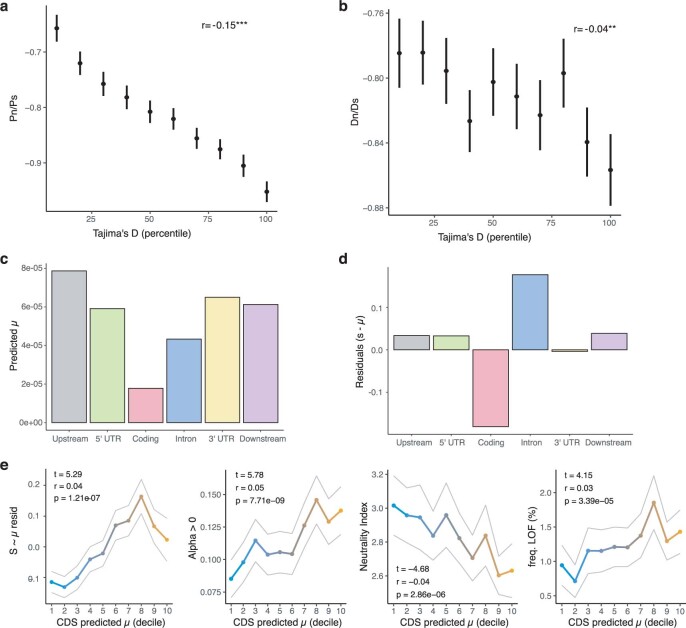
Extended Data Fig. 9. Predicted mutation rates and evidence of selection on natural polymorphisms versus de novo mutations across gene regions.
a, Relationship between Tajima’s D of gene bodies and coding region selection estimated by Pn/Ps (n = 21,407 genes) and b, Dn/Ds (n = 21,407 genes). c, Epigenome-predicted mutation probability in different gene features. d, Scaled residuals ((Obs-Pred)/Pred) from S ~ u. Significantly negative residuals in coding regions are consistent with purifying selection in natural populations acting on new mutations. e, Relationships between epigenome-predicted mutation probability and other estimates of constraint. Residuals between predicted mutation rate and observed mutations are positively correlated with predicted mutation rate indicating that genes subject to purifying selection are predicted to mutate less. Genes with low predicted mutation rates are also less likely to have alpha > 0, a measure of variants under positive selection. Genes with low predicted mutation rate are depleted in natural populations for non-synonymous variants that reach fixation, as measured by the Neutrality Index, and for loss-of-function variants.