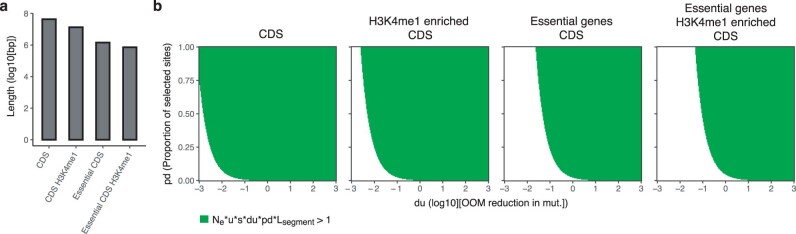
Extended Data Fig. 10. Estimates of Lsegment for different regions.
a, Length of sequence space (Lsegment) reflecting different types of regions. b, Test of parameter space that satisfies population genetic theoretical predictions for the possibility for targeted hypomutation to evolve. OOM = orders of magnitude. Selection on intragenomic mutation rate variation will be effective5,40 when Ne * u * s * du * pd * Lsegment > 1 where Ne is the effective population size, u is the mutation rate, s is the average selection coefficient on deleterious mutations, du is the degree of change in mutation rate, pd is the proportion of sites subject to purifying selection, and Lsegment is the region of the genome affected. Assuming an effective population size of ~300,000 (ref. 86–88), a mutation rate of ~10−8 (ref. 13), an average selection coefficient of 0.01 (ref. 5), an order-of-magnitude reduction in mutation rate5, and functionally constrained regions where 20% of sites are under selection5, the total length of the sequence affected, Lsegment, would have to be at least ~200 kb, which (accounting for differences in effective population size) is similar to previous estimates in humans5. For perspective, this minimum Lsegment is considerably shorter (~1.5%) than the sum of coding regions with elevated levels of H3K4me1 (top quartile is ~13 Mb, or 15% of the genome), a feature enriched in gene bodies and essential genes and associated with lower mutation rate. Thus, selection is expected to act with high efficiency on variants that cause DNA repair and protection mechanisms to preferentially target such regions.