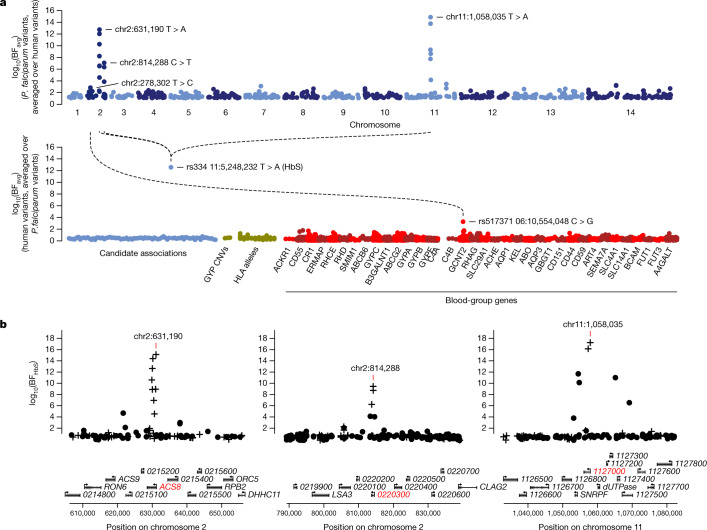
Fig. 1. Three regions of the P. falciparum genome are associated with HbS.
a, Points show the evidence for association between each P. falciparum variant and human genotypes (top row) or between each included human variant and P. falciparum genotypes (bottom row). Association evidence is summarized by averaging the evidence for pairwise association (Bayes factor (BF) for test in n = 3,346 samples) between each variant (points) and all variants in the other organism against which it was tested (log10 (BFavg)). P. falciparum variants are shown grouped by chromosome, and human variants are grouped by inclusion category as described in text and Methods. Dashed lines and variant annotations reflect pairwise tests with BF > 106; only the top signal in each association region pair is annotated (Methods). b, Detail of the association with HbS in the Pfsa1, Pfsa2 and Pfsa3 regions of the P. falciparum genome. Points show evidence for association with HbS (log10 (BFHbS)) for each regional variant. Variants that alter protein coding sequence are denoted by plus, and other variants are denoted by circles. Results are computed by logistic regression including an indicator of country as a covariate and assuming an additive model of association, with HbS genotypes based on imputation from genome-wide genotypes as previously described8. Mixed and missing P. falciparum genotype calls were excluded from the computation. Below, regional genes are annotated, with gene symbols given where the gene has an ascribed name in the PlasmoDB annotation (after removing ‘PF3D7_’ from the name where relevant); the three genes containing the most-associated variants are shown in red. A corresponding plot using directly typed HbS genotypes is presented in Extended Data Fig. 2.