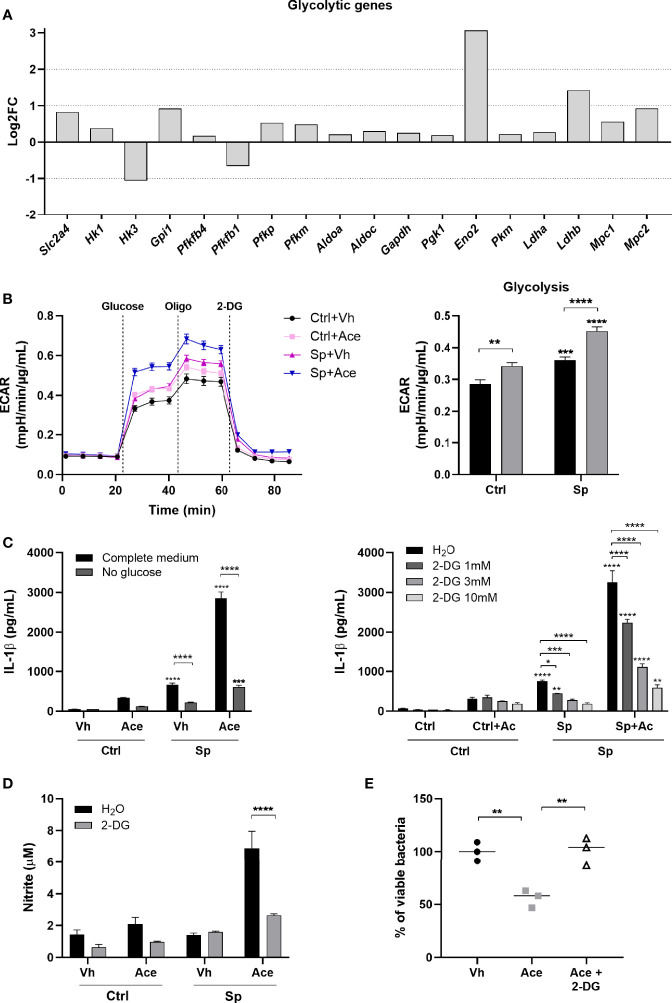
Figure 5.
Acetate enhances glycolysis leading to higher IL-1β production. (A) Log2 fold change of glycolytic genes expression of acetate treated MPI cells stimulated with S. pneumoniae over vehicle treated cells stimulated with S. pneumoniae. Data were obtained from RNA-Seq. All represented genes had p < 0.01 in the comparison between the two groups. Solute carrier family 2, facilitated glucose transporter member 4 (Slc2a4), hexokinase (Hk1, Hk3), glucose-6-phosphate isomerase (Gpi1), 6-phosphofructo-2-kinase (Pfkfb4, Pfkfb1), ATP-dependent 6-phosphofructokinase platelet and muscle types (Pfkp, Pfkm), fructose-bisphosphate aldolase (Aldoa, Aldoc), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), phosphoglycerate kinase 1 (Pgk1), gamma-enolase (Eno2), (Pkm), L-lactate dehydrogenase (Ldha, Ldhb), mitochondrial pyruvate carrier (Mpc1, Mpc2). (B) Glycolysis stress assay of MPI cells pre-treated or not with acetate, and then stimulated or not with S. pneumoniae for 24 h. ECAR was measured in Seahorse after injection of glucose, oligomycin (Oligo) and 2-Deoxy-D-glucose (2-DG) (right panel). Glycolysis was calculated by the subtraction of the highest measure after glucose injection by the last basal measure, for each group (left panel). (C) ELISA for IL-1β from supernatant of MPI cells pre-treated or not with acetate in the presence or not of glucose (left panel) or 2-DG (right panel) and then stimulated or not with S. pneumoniae for 24 h. (D) Nitrite concentration assessed by Griess assay from supernatant of MPI cells pre-treated or not with acetate in the presence of absence of 2-DG 10 mM and stimulated or not with S. pneumoniae during 48 h. (E) % of intracellular viable bacteria left 6 h post infection of activated MPI cells treated or not with acetate and/or 2-DG 10 mM, normalized to vehicle group. Lines/bars showing the mean and error showing the SEM of sextuplicates (B) or triplicates (C, D) and data showing the median of triplicates (E). Results are representative of three independent experiments. Statistical analysis was done using Two-way ANOVA (B, C) and One-Way ANOVA (E) corrected with Sidak’s multiple comparisons test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).