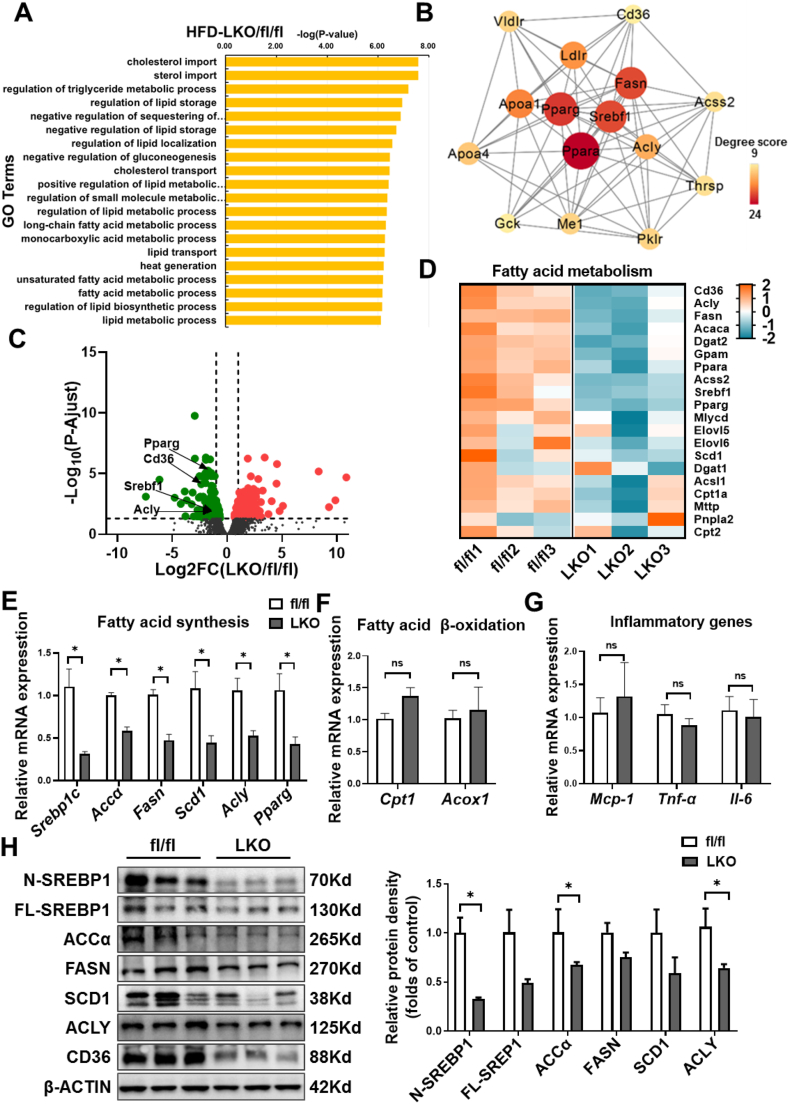
Figure 3.
Decreased hepatic lipogenesis in LKO mice. RNA sequence analysis of the livers from fl/fl and LKO mice under 16-week-HFD (n = 3). A: Top 20 biological process gene ontology (GO) terms enriched by differentially expressed genes (DEGs) in fl/fl and LKO mice. The DEGs were selected with a criterion at adjusted P-value < 0.05. B: The protein–protein interactions (PPI) network analysis of genes including in top 20 BP GO terms. The top 15 hub genes were selected and visualized with Cytoscape software. The size and color of map nodes were determined by degree score. C: Heatmap representation of genes involved in fatty acid metabolism. The gradual color ranged from pink to blue represented the mRNA expression changing from up-regulation to down-regulation. D: Volcano plot comparison of genes in fl/fl versus LKO mice, showing the distribution of significance [-log10 (P adjust value)] vs. fold change (FC) [log2 (FC)] for all genes. The red points represented up-regulated genes (log2 (FC) > 1 and P adjust value < 0.05, 222 genes) and green points represented down-regulated genes (log2 (FC) < −1 and P adjust value < 0.05, 222genes), while gray points indicated genes without significant differential expression (|log2 (FC) | < 1 or a P adjust value > 0.05). E–G: The liver mRNA levels of lipogenic genes (E), inflammatory genes (F), and fatty acid β-oxidation genes (G) determined by Q-PCR (n = 3). H: Hepatic expression of the SREBP1 and downstream lipogenic enzymes determined by immunoblotting. Quantitation of protein levels was shown in the right (n = 3). ∗P < 0.05 vs. fl/fl mice.