Figure 5. The structural basis for inhibition of GudB by GltA binding.
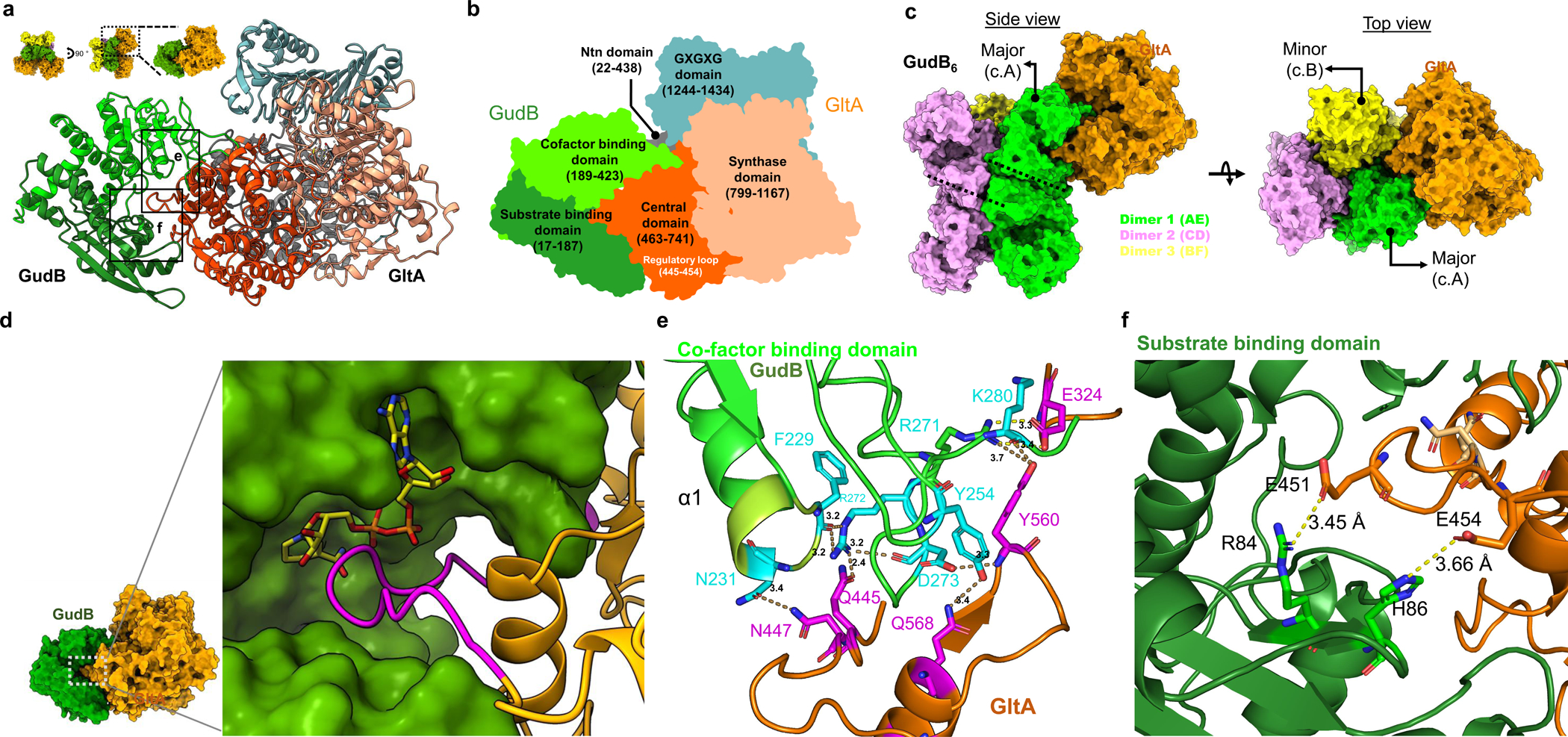
a) Zoom-in model of the GudB-GltA interaction. The colors represent the different domains of GltA and GudB as detailed in the next panel. The inset shows the perspective of the model with respect to the model shown in Fig. 4f. The dashed box (G) corresponds to the region containing residues from GudB and GltA forming a hydrogen bonding network. This is shown in detail in panel G. b) GltA consists of Ntn amidotransferase (Ntn) domain (grey), a central domain (orange), a synthase domain (sand) and a GXGXG domain (metal blue). The central domain of GltA (orange) interacts with residues from both the substrate (green) and co-factor binding domain of GudB (light green). c) GudB’s hexamer comprises a trimer of dimer; GltA (in orange) interacts with two protomers of GudB (major and minor) from two different dimers (green and yellow, respectively). The front view shows the major interacting GudB protamer, and the top view the minor one. The coloring pattern of the three dimers and the corresponding chain ID’s are also shown. The dimer interface in two of the dimers (1 and 2) is shown as dashed lines. d) Zoom-in on GudB active site cleft (surface display, in green) and the interacting loop of GltA (magenta; the remaining structure of GltA is in orange). NAD+ (in yellow sticks) is modelled into the active site cleft of GudB based on superposition of GltA-bound GudB to 1V9L (open state). The steric overlap of GltA’s loop with NAD+ would be even higher in the closed state of the enzyme. e) Zoom-in on the boxed region shown in panel A, depicting the hydrogen bonding network between residues of GltA (magenta) and the co-factor binding domain of GudB (cyan). Notably, GudB’s N231, which is located at the tip of the phosphate binding loop (and binds the NAD+’s phosphate groups) is bound to GltA’s N447, and thus directly interferes with NAD+ binding. f) The interaction of GltA’s E451 and E454 with residues of the substrate binding domain of GudB (R84 and H86).
