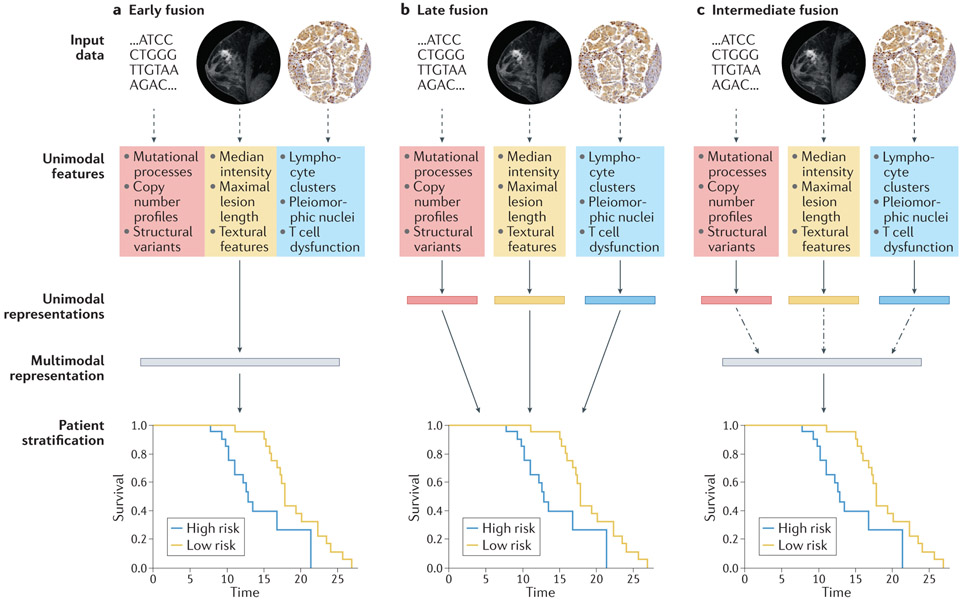
Figure 3. Design choices for multimodal models with genomic, radiological, and histopathological data.
Solid arrows indicate stages with learnable parameters (linear or otherwise), dashed arrows indicate stages with no learnable parameters, and dashed and dotted arrows indicate the option for learnable parameters, depending on model architecture. (a) In early fusion, features from disparate modalities are simply concatenated at the outset. (b) In late fusion, each set of unimodal features is separately and fully processed to generate a unimodal score before amalgamation by a classifier or simple arithmetic. (c) In intermediate fusion, unimodal features are initially processed separately prior to a fusion step, which may or may not have learnable parameters, and subsequent analysis of the fused representation. All schemata shown are for deep learning (DL) on engineered features: for convolutional neural networks (CNNs) directly on images, unimodal features and unimodal representations are synonymous. For linear machine learning (ML) on engineered features, no representations are learned between features and stratification. The magnetic resonance imaging (MRI) images were obtained from The Cancer Imaging Archive (TCIA). Histology images were obtained from the Stanford Tissue Microarray database, ref. 155.