Figure 5.
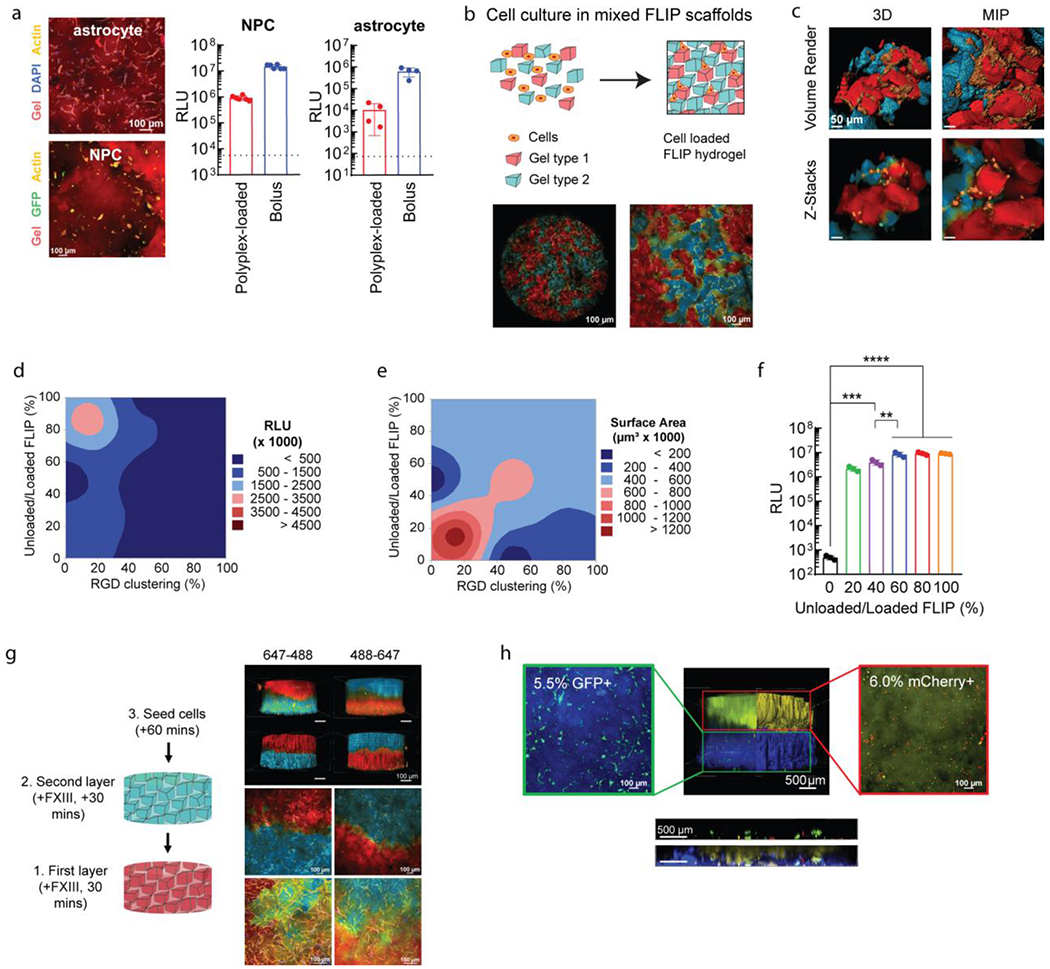
Further applications of FLIP scaffolds for plasmid transfection. (a) Transfection of relevant CNS cell types in 3D culture. Confocal images of stained cell cultures for human astrocytes and mouse-derived primary neural progenitor cells (NPCs). Cells were transfected using HA-NB coated DNA/PEI particles in FLIP (n=4), compared to bolus transfection and the cell control (dotted line), after 48 hours. (b) Altering FLIP properties from sHMP mixing, with confocal imaging for MSCs seeded within mixed colored FLIP scaffolds. Cell spreading within FLIP scaffolds comprised of two different precursor solutions, stained for 488 and 647, with cell membrane staining of live cells. Note that the 488 channel is pseudocolored blue, and the 647 channel red. (c) High-resolution confocal imaging of the mixed scaffold, showing the interface of cells spreading between the two types of sHMP. Both z-stacks and volume renderings are shown, from a 3D orthogonal view and from maximum-intensity projection (MIP). (d-e) DOE optimization of cell transfection (d) and spreading or surface area (e) based on RGD peptide clustering on sHMP and ratio of nanoparticle-loaded to non-loaded sHMP within FLIP scaffolds. Transfection was assessed with GLuc. RGD clustering corresponds to the ratio of gel exposed to reactive RGD peptide at the time of precursor formulation, with a fixed 1 mM concentration. 100% modified RGD-modified HA meant that the scaffold was homogenous (i.e. the entire HA-AC precursor was reacted with 1 mM RGD), while 10% modified and 90% non-modified meant that there was 10% clustering on the microgels, and 0% referred to a gel without any RGD (0 mM). Surface area was assessed based on confocal imaging of the 3D cultures and rending the z-stacks in IMARIS for volume filling and surface area measurements from actin labeling. (f) An example of the transgene output is shown in for a fixed 20% RGD clustering and was determined that 60-80% nanoparticle content and 20% RGD clustering was the optimal balance between transfection and spreading/viability of cells. A one-way ANVOA with Tukey HSD was performed on the samples (n=3), with significance reported at p <0.05 (*), <0.01 (**), <0.005 (***), and <0.001 (****). (g) FLIP gel layering based on sequencing syringe loading and injection into the 3D culture mold, with the ability to alternate layers of 488 or 647 stained gels without merging or loss of gels due to the annealing chemistry. Again, the 488 channel is pseudocolored blue, and the 647 channel red. 4X images were processed for volume rendering to show the entire scaffold distribution. 10X and 20X images were taken at the interface of the scaffold layers to demonstrate cell infiltration across the layers, shown as MIPs. (h) Application of FLIP scaffolds for “domain transfection,” in which different gel layers contained DNA/PEI particles with transgenes for either GFP (AF305-labelled layer, blue) or mCherry (AF647-labelled layer, yellow). Transgene expression was assessed with flow cytometry on degraded gels for cell extraction (separated by layer), showing highly specific gene expression for only the given reporter in each layer (denoted in the microscopy image of reporter-expressing cells by the percentage). The FLIP scaffolds were also imaged for z-stacks at 4X across the layer interface to assess transfection of GFP and mCherry in the region, shown with the horizontal ortho-projection, both with and without the gel channels present.
