FIGURE 3.
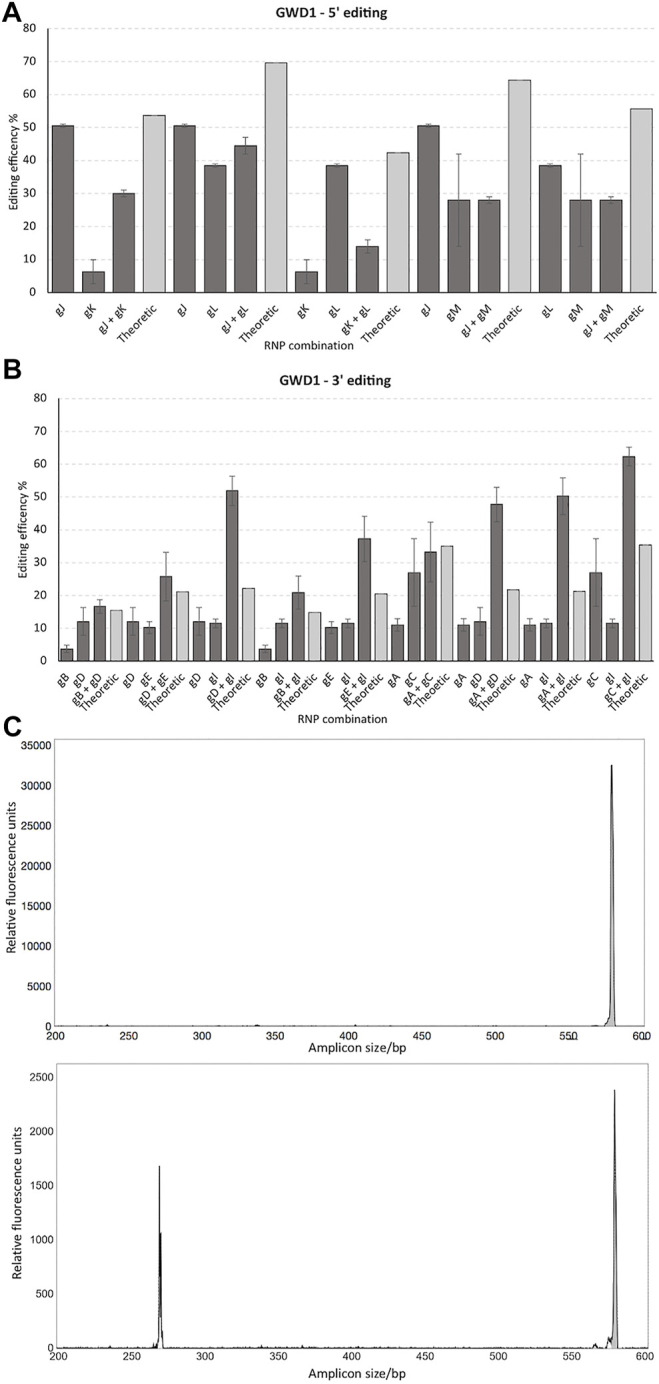
Targeting the 5′ versus the 3′ end of GWD1 and effect of multiplexing. 4 and 6 gRNAs were designed for the 5’ (gJ, gK, gL, and gM) (A) and 3’ (gA, gB, gC, gD, gE, and gI) (B) end ofGWD1, respectively. The gRNAs were tested individually and in combination. Theoretical designates the probability of editing, calculated as the multiplied probabilities of each gRNA not conferring editing, (1 – gRNA1 editing fraction) * (1 – gRNA2 editing fraction) subtracted from 1, i.e., 1 – [(1 – gRNA1 editing fraction) * (1 – gRNA2 editing fraction)] * 100 (%), where 1 designates all alleles in the cell pool. (C): IDAA chromatogram of WT amplicon (upper panel, grey) and gD and gI combined editing (lower panel) including the unedited WT amplicon (grey) andminor editing from either gDor gI and where the lower sized amplicons include the combined editing of gD and gI andthus the gD and gI delineated deletion. The X-axis and Y-axis show the size of the amplicons in bp and relative fluorescence, respectively. IDAA primer pairs used for scoring gA, gB, gC, gD, gE & gI: GWD Forward primer 1 + GWDReverse primer 1 and for scoring gJ, gK, gL & gM: GWD forward primer 2 + GWD reverse primer 2 (see Materials and Methods).
