Fig. 3: Re-reading of a single peptide.
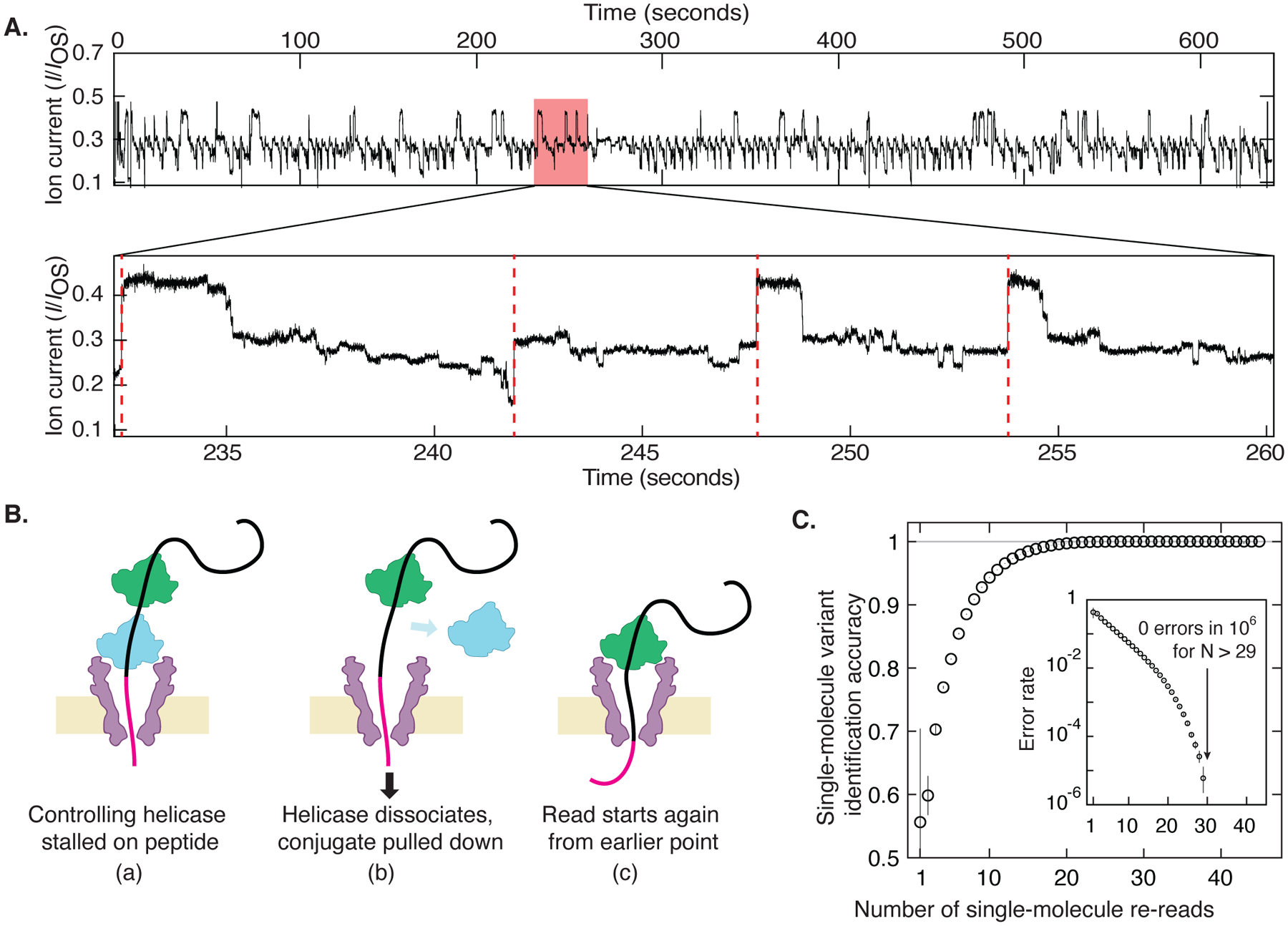
(A) Highly repetitive ion current signal corresponding to numerous re-reads of the same section of an individual peptide (in this case, the G-substituted variant). The expanded plot below shows a region that contains four rewinding events (red dashed lines), where the trace jumps back to level 52 ± 2 of the consensus displayed in Fig. 2A. (B) Re-reading is facilitated by helicase queueing, where (a) a second helicase binds behind the primary helicase that controls the DNA-peptide conjugate, re-reading starts when (b) the primary helicase dissociates, and (c) the secondary one becomes the primary helicase that drives a new round of reading. (C) By using information from multiple re-reads of the same peptide, the identification accuracy can be raised to very high levels of fidelity. These results indicate that with sufficient numbers of re-reads, random error can be eliminated and single-molecule error rate can be pushed lower than 1 in 106 even with poor single-pass accuracy. Inset is a logarithmic plot of the error rate = 1 - accuracy.
