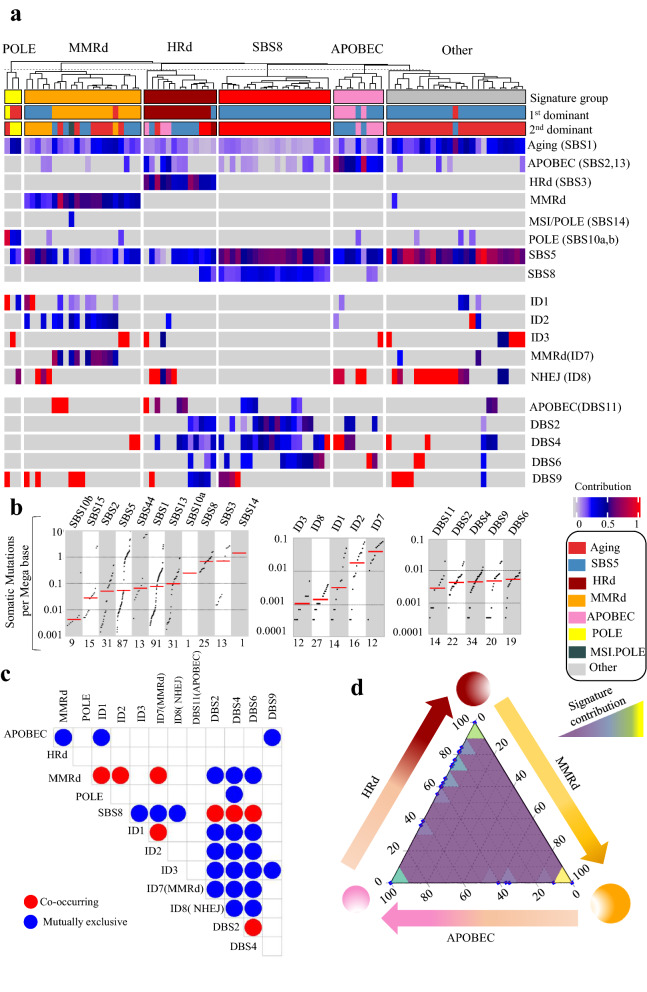
Fig. 1.
Mutational signature analysis of UCEC WGS tumors. a NMF-based de novo Mutational signatures of UCEC tumors visualized by a heatmap divided based on distinct signature status. The first and second dominant signatures are annotated in the top. The dominant signatures were significantly associated with their corresponding signature group (HRd: P 4.47E-12, MMRd: P < 2.2e-16, POLE: P 8.23E-06, APOBEC: P 2.81E-10 and SBS8 P < 2.2e-16; Fisher exact test). The contribution values of each signature are shown by a color scale. Color codes representing each dominant mutational signature are shown. b TMB of SBS, ID and DBS signatures for UCEC tumors. TMB is measured in somatic mutations per Megabase (Mb). In the TMB plots, columns represent the detected mutational signatures and are ordered by mean somatic mutations per Mb from the lowest frequency, left, to the highest frequency, right. Numbers at the bottom of the TMB plots represent the numbers of tumors harboring each mutational signature. Only samples with counts more than zero are shown. c Interaction of signatures with each other. Also see Additional file 3: Table S2 for P values calculated by Hypergeometric test. d Ternary relation between MMRd, APOBEC and HRd mutational signatures. The ternary plot depicts the contribution of these three signatures as positions in an equilateral triangle. The contribution values of each signature are shown by a color scale. The size of circles represents the prevalence of each signature