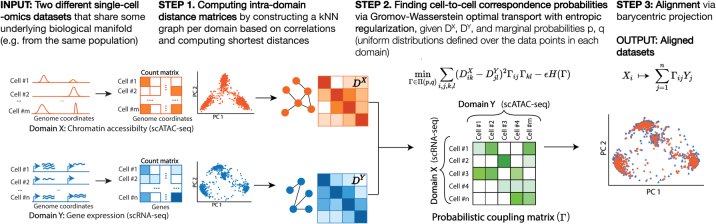
FIG. 1.
Overview of the SCOT algorithm: SCOT takes in two data sets of single-cell genomics measurements in the form of count matrices to align them. For each data set, it first constructs k-nearest neighbor graphs based on correlations between samples and calculates the distance matrices capturing the intra-domain distances. Given these, it solves the Gromov-Wasserstein optimal transport formulation to find an ideal coupling (also known as “correspondence”) matrix, describing the probability of alignment between the samples across the two data sets. Finally, it completes the alignment by projecting the first domain onto the second one based on correspondence probabilities using barycentric projection. SCOT, single-cell alignment using optimal transport.