ABSTRACT
Colorectal adenocarcinoma (COAD) is a prevalent malignant tumor. Cancer-associated fibroblasts (CAFs)-derived extracellular vesicles (EVs) (CAFs-EVs) are implicated in COAD treatment. This study explored the mechanism of CAFs-EVs in COAD. CAFs and normal fibroblast (NFs) were isolated from COAD tissues and adjacent normal tissues. Vimentin, α-SMA, and FAP expressions were detected. EVs were isolated from CAFs and identified. SW480 and HCT116 cells were co-incubated with EVs. The EV uptake and COAD cell malignant behaviors were assessed. EV-treated SW480 and HCT116 cells were co-cultured with human umbilical vein endothelial cells (HUVECs). Extensive analyses were conducted to examine HUVEC proliferation, migration, and angiogenesis, and miR-135b-5p expression in COAD cells, and SW480 and HCT116 cells. CAFs were transfected with the miR-135b-5p inhibitor. miR-135b-5p downstream targets were predicted. FOXO1 expression in the co-culture system was determined and then overexpressed to evaluate its role in HUVECs mediated by COAD cells. COAD mouse model was established by transplanting SW480 cells into nude mice and injecting with EVs. Tumor growth rate, volume, and weight were examined. Ki67, VEGF, CD34, FOXO1 expressions, and VEGF content were detected. CAFs-EVs promoted COAD cell malignant behaviors and COAD cells-mediated HUVEC proliferation, migration, and angiogenesis. CAFs-EVs delivered miR-135b-5p into COAD cells. miR-135b-5p targeted FOXO1. Inhibition of miR-135b-5p in EVs or overexpression of FOXO1 partially reversed the effect of EVs on promoting COAD-induced angiogenesis. CAFs-EVs promoted tumor proliferation and angiogenesis of COAD in vivo. CAFs-EVs delivered miR-135b-5p into COAD cells to downregulate FOXO1 and promote HUVECs proliferation, migration, and angiogenesis.
KEYWORDS: Colorectal adenocarcinoma, Cancer-associated fibroblasts, extracellular vesicles, angiogenesis, miR-135b-5p, FOXO1, SW480/HCT116, HUVECS
Introduction
Colorectal adenocarcinoma (COAD) has persisted as the most common type of cancer worldwide with definitive characteristics of dysregulated intestinal epithelial differentiation, apoptosis, and proliferation.1,2 Statistically, it is regarded as the 3rd in the global cancer incidence rate and the 4th in global cancer associated mortality accounting for over 1 million newly diagnosed cases and about 700000 deaths annually.3 Notably, several risk factors for COAD have been identified which include smoking, excessive drinking, high consumption of processed and red meats, diabetes, obesity, inflammatory bowel disease, gender, age, and a family history of COAD.4 Currently, there are several therapeutic options available for COAD, such as anti-angiogenic agents, anti-epidermal growth factor receptor, and combination chemotherapy, however none of them can completely cure COAD, and thus further investigations are warranted for complete treatment of COAD.1
Cancer-associated fibroblasts (CAFs) are the primary constituents of tumor stroma as evidenced by a highly activated myofibroblastic phenotype with an increased proliferation rate, which are the most vital components of the tumor microenvironment.5 CAFs are essential regulators in the initiation and development of numerous types of cancers corresponding to their ability to produce terminal growth factors, proteases, and cytokines, thus fundamentally manipulating the cell motility, proliferation, inflammatory responses, along with the deposition and remodeling of extracellular matrix.6 CAFs are associated with increased cancer angiogenesis and the development of COAD.7 An existing study identified that CAFs and cancer cells can release extracellular vesicles (EVs) to manipulate the effects of each other.8 EVs are small vesicles with a diameter of 30–150 nm that can transfer the biological macromolecules of RNAs, proteins, and lipids.9 EVs can notably influence the regulation of malignant cell behaviors using cargo molecules and are evidently implicated in a variety of cancers.10 CAFs can improve chemoresistance and stemness of COAD by the transfer of EVs-derived lncRNA H19.11 As CAFs-EVs elicited the characteristics of tumor stimulation, hence they are associated with a low survival rate in various cancers, thus demonstrating functionality as potential therapeutic targets.8 Currently, limited studies have investigated the function of CAFs-derived EVs (CAFs-EVs) in the angiogenesis in COAD. The effects of CAFs-EVs on COAD and its mechanism warrant extensive investigation.
EVs are vital for intercellular communication as they can essentially deliver microRNAs (miRNAs) into the tumor cells.12 Moreover, miRNAs have been identified as chief regulators for COAD.13 An existing study identified that miR-135b-5p can promote the angiogenesis and proliferation of endothelial cells in diabetic retinopathy mice.14 Upregulation of miR-135b-5p can radically enhance the pro-angiogenic ability of human umbilical cord mesenchymal stem cells (hUC-MSC) derived EVs.15 Previously, miR-135b-5p has been identified to be upregulated in COAD tissues.16 However, whether the CAFs-EVs can carry miR-135b-5p into COAD cells and function in the angiogenesis of COAD is largely unknown. We predicted the downstream target genes of miR-135b-5p using a combination of bioinformatics analysis and screening, after which the forkhead box protein O1 (FOXO1) was identified. FOXO1 is one of the forkhead transcription factors, which functions as an essential regulator for angiogenesis.17 Notably, the knockdown of miR-135b can increase FOXO1 in colorectal cancer.18 Presently, no study has investigated the effect and mechanism of CAFs-EVs-derived miR-135b-5p on COAD and whether the CAFs-EVs can influence COAD through miR-135b-5p/FOXO1. The current study aims to investigate the mechanism of CAFs-EVs in regulating COAD.
Materials and methods
Ethics statement
The experiments were conducted with approval of the academic ethics committee of HwaMei Hospital, University of Chinese Academy of Sciences. All procedures were in compliance with the code of ethics. Significant measures were taken to minimize the number of animals used and their suffering.
Isolation, culture, and identification of CAFs
Fresh tissue samples and adjacent normal tissues of COAD were collected from patients after operations in HwaMei Hospital, University of Chinese Academy of Sciences. A portion of the tissue was immediately reserved for isolation and culture CAFs of colorectal cancer tissue. CAFs or normal fibroblast (NFs) were isolated in strict accordance with an existing protocol.5 Firstly, the colon tissue was divided into small sections, vibrated and detached using Hank’s balanced salt solution (HBSS) containing a combination of 1 mg/mL collagenase, 2.5 mg/mL trypsin, and 30 μg/mL DNase I in a 37°C water bath. The detachment was performed for 3 cycles of 30 min each. The detached mixed cells were cultured with 10% fetal bovine serum-Dulbecco’s modified Eagle’s medium (FBS-DMEM). After the third passage, the cells formed a monolayer with identical morphology as the fibroblast-like cells. An analysis revealed that the samples contained more than 90% of vimentin-positive cells. The isolated NFs and CAFs were cultured in 10% FBS-DMEM for 24 h. After intensive fluorescent staining, the samples were co-incubated with the primary vimentin antibody (ab92547, 1/250, Abcam, Cambridge, MA, USA), smooth muscle alpha-actin (α-SMA) antibody (ab7817, 5 μg/mL, Abcam), and fibroblast activation protein (FAP) antibody (ab207178, 1/100, Abcam) respectively. Next, the samples were supplemented with a secondary antibody. Immunofluorescence was observed under a fluorescence microscope (Olympus Life-Sciences, Tokyo, Japan).
Subsequently, the purity of CAFs was detected by means of flow cytometry. CAFs were detached into single cells with 0.25% trypsin with termination of the detachment using 10% FBS-DMEM. The samples were subject to centrifugation at 150 g for 6 min. After resuspension with the FACS buffer (100 μL/106 cells, Flow Cytometry Staining Buffer, phosphate buffer saline (PBS) (without calcium and magnesium ion in the solution) + 1% FBS), the samples were supplemented with PE anti-human FAP antibody (ab207178, 10 μL/106 cells), incubated at 4°C in conditions devoid of light for 30 min, and evaluated on the machine to calculate the proportion of FAP-positive cells.
Isolation, identification, and grouping of CAFs-EVs and NFs-EVs
The EVs in FBS were eliminated by ultra-centrifugation of the culture medium at 100000 g at 4°C overnight. Upon attaining 80% confluence, the supernatant was removed from CAFs or NFs. Subsequently, the samples were supplemented with 10% EV-free FBS (EBM-2, Lonza, Allendale, NJ, USA) and cultured in a CO2 incubator at 37°C for 48 h. The collected supernatant was subject to centrifugation at different speeds. The procedure steps were as follows: at 300 g at 4°C for 10 min, at 2000 g at 4°C for 15 min, at 5000 g at 4°C for 15 min, at 12000 g at 4°C for 30 min, and at 100000 g at 4°C for 70 min. The sample was suspended with PBS after removal of the supernatant. The sample was centrifuged at 100000 g at 4°C for 70 min with removal of the supernatant along with suspension of the sample in PBS prior to preservation at −80°C.
The isolated EVs were identified in strict accordance with the following protocols: the morphology of isolated EVs was observed under transmission electron microscopy (TEM); the size distribution of EVs was analyzed using the nanoparticle tracking analysis (NTA); Western blot (WB) was adopted to verify the expression patterns of EV surface antigens CD9 (EBM-2, Lonza, Allendale, NJ, USA), CD63 (ab271286, Abcam), and calnexin (ab22595, Abcam), and the supernatant of NFs or CAFs supplemented with GW4869 was used as negative control (NC).19 The identified EVs were lysed with the lysate, while the protein quantification was conducted using the bicinchoninic acid (BCA) protein quantitative kit (Boster, Wuhan, Hubei, China) according to the provided instructions. The protein content was used as the standard of EV concentration.
The EVs used in this study were assigned into the following three groups: the EVs group, the EVs-miR-NC group (EVs extracted from CAFs transfected with inhibitor NC), the EVs-miR-inhi group (EVs extracted from CAFs transfected with miR-135b-5p inhibitor). The method of miRNA transfection was as follows: CAFs in the logarithmic growth phase were collected and seeded in 6-well plates at a concentration of 1.0 × 105 cells/well. Upon attaining 60% cell confluence overnight, the miR-135b-5p inhibitor or the corresponding NC (unrelated sequence NC) was transfected into the adherent cells at a concentration of 50 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in strict accordance with the provided instructions. After 48 h of transfection, the cells were collected for subsequent experimentation. The miR-135b-5p inhibitor and unrelated sequence NC were synthesized and purified by GenePharma (Shanghai, China).
Cell culture and grouping
Human COAD cell lines SW480 (CCL-228) and HCT116 (CCL-247) and human umbilical vein endothelial cells (HUVECs) were provided by American Type Culture Collection (ATCC, Manassas, VA, USA) for this experiment. The cells were cultured in McCoy’s 5A complete medium or eibovitz’s L-15 complete medium containing a combination of 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. All experiments were strictly conducted in cells between the 10th and 20th generations. The cells were cultured in a humidified incubator at 37°C containing 95% O2 and 5% CO2.
SW480/HCT116 cells were assigned into the following 6 groups: the NFs-EVs group (20 μg NFs-EVs were added for co-incubation), the CAFs-EVs group (20 μg EVs was added for co-incubation), the EVs-miR-NC group (20 μg EVs-miR-NC was added for co-incubation), the EVs-miR-inhi group (20 μg EVs-miR-inhi was added for co-incubation), the oe-FOXO1 + EVs (the cells were treated with 20 μg EVs after FOXO1 overexpression), and the oe-NC + EVs group (the cells were treated with 20 μg EVs after transfection with oe-FOXO1 negative control oe-NC).
EVs uptake assay
SW480/HCT116 cells were stained using the CellTrace CFSE cell proliferation kit (Invitrogen) in strict compliance with the provided instructions. The EVs were labeled using the PKH26 Fluorescent Cell Linker Kit (Sigma-Aldrich, St. Louis, MO, USA). The EVs with 20 μg protein content were mixed with 1 mL PKH26 dye solution (at a dilution ratio of 1:1000) for 20 min. The samples were rinsed with PBS prior to centrifugation at 1000000 g for 70 min. Next, the PKH26-labeled EVs were supplemented into CFSE-labeled cells for 24 h of co-culture. The uptake of EVs at different time points was observed under the laser confocal fluorescence microscope (TCS SP8, Leica, Wetzlar, Germany).
Cell counting kit-8 (CCK-8)
Cell proliferation was measured by CCK-8 (Dojindo Laboratories, Kumamoto, Japan) in strict compliance with the provided instructions. In order to measure the cell proliferation, 100 μL HUVECs were dripped into 96-well plates (2 × 103 cells/well) prior to incubation for 24 h. The supernatant of tumor cells from each group was collected and added into the corresponding wells at the ratio of 0%, 20%, 40%, 60%, 80%, and 100% for CCK-8 detection. The absorbance value was detected at the excitation wavelength of 450 nm using the microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Transwell assays
Transwell assay was performed to detect the concentration of SW480/HCT116 COAD cells. A total of 5 × 104 cells from different treatment groups were immersed in 200 μL serum-free medium and added into the Transwell plate coated or uncoated with Matrigel (BD Biosciences, San Jose, CA, USA). The basolateral chamber was supplemented with 600 μL of 10% FBS. After 24 h of incubation, the samples were fixed with methanol for 20 min, stained with 0.1% crystal violet solution, and analyzed under a microscope.
Transwell assay was performed to detect concentration of HUVECs co-cultured with the SW480/HCT116 cells. The tumor cells in each group (4 × 105 cells/well) were immersed in 500 μL culture medium and supplemented into the basolateral chamber of the Transwell plate without Matrigel coating for overnight incubation. The apical chamber was paved with HUVECs (1 × 105 cells/well) and supplemented with 200 μL of FBS-free medium. The samples were incubated at 37°C for 6 h and the observations of migrated cells were documented and counted under a microscope.
Tube formation assay
The basolateral chamber was supplemented with 200 μL of Matrigel. The samples were incubated at 37°C for 30 min. HUVECs were seeded in 24-well plates (2 × 105 cells/well). The transfected cells (4 × 105 cells/well) were placed in an apical chamber supplemented with 200 μL of medium containing FBS. The samples were incubated at 37°C for 6 h. The formed tubes were counted and documented under a microscope.
Enzyme-linked immunosorbent assay (ELISA)
The supernatant of HUVECs co-cultured with SW480/HCT116 cells was isolated for subsequent experimentation. ELISA kit (R&D Systems) was adopted to measure the concentration of vascular endothelial growth factor (VEGF). The absorbance value at OD450 wavelength was measured using a microplate reader.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract the total RNA content. The PrimeScript RT reagent kit (Takara, Dalian, China) was used for reverse transcription of the RNA content into cDNA. The qPCR was performed in strict compliance with the ABI PRISM 7900 sequence detection system of SYBR Green II (Takara Biotechnology). The reaction conditions were as follows: pre-denaturation at 95°C for 5 min and 40 cycles of denaturation at 95°C for 15s, annealing at 60°C for 20 s and extension at 72°C for 35s. GAPDH and U6 were regarded as internal parameters while the analysis was conducted based on the 2−ΔΔCt method. The primer sequences synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) are shown in Table 1.
Table 1.
Primer sequence
| Gene | Forward 5ʹ-3’ | Reverse 5ʹ-3’ |
|---|---|---|
| miR-135b-5p | AGGGCACAGGAGGGGC | AGTGCAGGGTCCGAGGTATT |
| FOXO1 | CGTAAGCTGGGAGAGACCTG | GGATGGATGGGAATGAACAC |
| GAPDH | CTCAGACACCATGGGGAAGGTGA | ATGATCTTGAGGCTGTTGTCATA |
| U6 | ATTGGAACGATACAGAGAAGATT | GGAACGCTTCACGAATTTC |
Tumor model establishment
BALB/C female athymic nude mice (6 weeks old) were selected for establishment of the COAD mouse model. The mice were provided by Beijing Vital River Laboratory Animal Technology Co., Ltd. [SCXK (Beijing) 2019–0009, Beijing, China]. Each nude mouse was subcutaneously injected with 0.2 mL of SW480 mixed with about 1 × 107 cells. The different treatment groups were injected with 100 μg of EVs (50 μg/100 μL PBS) via the tail vein once every 4 day for a total of 28 days. The length (L) and width (W) of the tumor were measured daily using a vernier caliper. The tumor volume was calculated based on the formula (length × width2)/2.
The nude mouse model of COAD used in this study was assigned into the following three groups: COAD + EVs (EVs were injected through the tail vein), COAD + CAFs-EVs-miR-NC (CAFs-EVs-miR-NC was injected through the tail vein), and COAD + CAFs-EVs-miR-inhi (CAFs-EVs-miR-inhi was injected through the tail vein). In this study, a total of 36 nude mice were chosen for COAD model establishment. Each group had 12 mice, with 6 reserved for observation and measurement of the tumor growth rate, volume, and weight on the 7th, 14th, 21st, and 28th day, and the tumor tissues on the 28th day were sectioned for immunohistochemistry detection of Ki67, VEGF and CD34 expressions, while the remaining 6 mice were reserved for RT-qPCR detection of miR-135b-5p and FOXO1 mRNA expressions.
Immunohistochemistry
Tumor sections were prepared for immunohistochemical analysis. The samples were paraffin-embedded and sectioned. After a regimen of dewaxing, dehydration, inactivation, and blocking, the samples were incubated with CD34 (ab81289, at a dilution ratio of 1:2500, Abcam), Ki67 (ab15580, 5 µg/mL, Abcam) antibodies or VEGF (ab52917, at a dilution ratio of 1:100, Abcam) antibody at 4°C overnight. The samples were incubated with a secondary antibody and subsequently stained with diazine. The cytoplasmic staining of the vascular endothelial cells was positive.
Western blot (WB)
Mouse tumor tissue homogenate was lysed with the enhanced radio-immunoprecipitation assay (RIPA) lysate (Boster) containing a protease inhibitor for 20 min and centrifuged at 4°C at 3000 g for 20 min and isolation of the supernatant. The protein concentration in the supernatant was detected using the BCA protein quantitative kit. The protein content was isolated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The isolated protein content was transferred onto polyvinylidene fluoride (PVDF) membranes. A membrane blockade was induced using 5% bovine serum albumin (BSA) at room temperature for 2 h to block any nonspecific binding, prior to incubation with the primary antibody FOXO1 (ab179450, Abcam) at 4°C overnight. After a rinse with the tris buffered saline-Tween20 (TBST), the sample was supplemented with horseradish peroxidase (HRP) labeled secondary antibody and incubated for 1 h at room temperature. The enhanced chemiluminescence (ECL) reagent (EMD Millipore, Bedford, MA, USA) was added for development. Image Pro Plus 6.0 (Media Cybernetics Inc., Silver Springs, MD, USA) was used to quantify the gray scale of bands in each group in WB images, with β-actin serving as an internal parameter. Each experiment was conducted three times independently.
Bioinformatics analysis
The downstream target genes of miRNA were predicted by a combination of the StarBase (http://starbase.sysu.edu.cn/), RNAInter (http://www.rna-society.org/rnainter/), Targetscan (http://www.targetscan.org/vert_71/) and miRDB (http://www.mirdb.org/). The coexpedia database was used to identify the co-expression relationship of genes and obtain the co-expression score of the website for subsequent screening of the target genes.
Statistical analysis
GraphPad Prism 8.01 (GraphPad Software Inc., San Diego, CA, USA) and SPSS 21.0 (IBM Corp. Armonk, NY, USA) were used for elaborate data analysis and mapping. Kolmogorov-SmiRnov test showed that the continuous variable was in normal distribution and expressed as mean ± standard deviation. The independent t test was used for comparison between two groups and one-way analysis of variance (ANOVA) was used for comparison among multiple groups. Tukey’s multiple comparisons test was adopted for the post hoc test. In all statistical references, a value of P < .05 was indicative of statistical significance.
Results
Isolation and identification of CAFs and their EVs
The CAFs and NFs from the COAD tissues and adjacent normal tissues of the patients undergoing COAD surgery were successfully isolated. NFs and CAFs elicited fibroblasts characteristics of long fusiform morphology, elevated expression pattern of the fibroblast marker vimentin, positive expression pattern of α-SMA, and positive expression pattern of fibroblast activating protein (FAP); compared with NFs, CAFs elicited enhanced vimentin, α-SMA and FAP fluorescence expression (Figure 1a). Flow cytometry demonstrated that more than 90% of the isolated cells were fibroblasts (Figure 1b), thus indicative of accurate CAF and NF isolation through separation and purification. Subsequently, the EVs and NFs were isolated from CAFs by ultracentrifugation. The EVs were in a circular or oval shape as revealed TEM with a diameter of 60–160 nm, with identification of intact membrane structure in the periphery of the EVs (Figure 1c). NTA was used for subsequent analysis to determine their size distribution. As demonstrated in Figure 1d, the predominant distribution of EVs was at about 100 nm, the concentration of NFs was 1.0 × 106 cells/mL, and the concentration of CAFs was 1.5 × 106/mL. WB analysis showed that elevated expression patterns of EV markers CD9 and CD63 in EVs, while no significant expression pattern was identified for Calnexin (Figure 1e). The preceding results are indicative of correct NFs-EVs and CAFs-EVs isolation.
Figure 1.
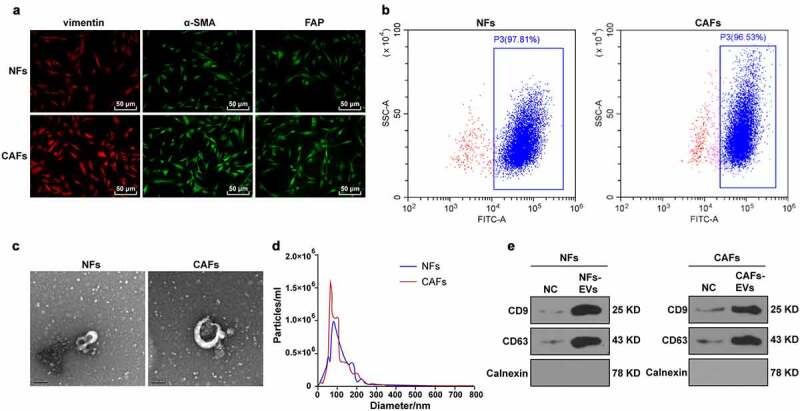
Identification of CAFs and EVs. The CAFs and NFs were successfully isolated from COAD tissues and adjacent normal tissues of patients undergoing colon cancer surgery. (a) Immunofluorescence was used to observe the morphology of CAFs and the expressions of surface markers vimentin, α-SMA, and FAP; (b) The purity of NFs and CAFs was detected using flow cytometry. The EVs were extracted. (c) The morphology of EVs was observed by TEM; (d) The size distribution of EVs was analyzed using NTA; (e) The expressions of CD9, CD63, and Calnexin were analyzed using WB.
CAFs-EVs promoted angiogenesis induced by COAD cells
Two COAD cell lines (SW480 and HCT116) were cultured and co-incubated with CAFs-EVs and NFs-EVs for 24 h. The results of the uptake test revealed that both of the EVs could be internalized by SW480/HCT116 cells (Figure 2a). To investigate the role of NFs-EVs or CAFs-EVs in the angiogenesis of COAD, the supernatants of SW480 and HCT116 cells treated with EVs were collected and co-cultured at variable ratios of 0%, 20%, 40%, 60%, 80%, and 100% with HUVECs. Cell proliferation was detected by the CCK-8 assay, which illustrated significantly increased proliferation of HUVECS after the addition of 60% of the supernatant of COAD cells treated with CAFs-EVs into the co-culture system (all P < .05), while NFs-EVs treated COAD cell supernatant had no significant effects on HUVEC proliferation (Figure 2b), thus indicating that CAFs-EVs could improve the proliferation of HUVECs mediated by COAD cells. Transwell assay demonstrated that compared with NFs-EVs treatment, CAFs-EV treatment could considerablyincrease the migration of HUVECs mediated by COAD cells (all P < .01) (Figure 2c). Furthermore, the number of connections was measured to quantify the degree of tube formation of HUVECs. After co-culture with CAFs-EVs-treated COAD cells, the angiogenesis ability of HUVECs was significantly enhanced. In comparison with the NFs-EVs group, the number of connections was significantly increased (all P < .01) (Figure 2d). ELISA was adopted to detect the content of VEGF in the HUVECs supernatant in each group, which revealed that the VEGF content in the supernatant of HUVECs co-cultured with CAFs-EVs-treated COAD cells was significantly higher compared to the NFs-EVs group (all P < .01) (Figure 2e). The preceding results indicated that CAFs-EVs had radically promoted the proliferation and migration of HUVECs mediated by COAD cells and promoted angiogenesis.
Figure 2.
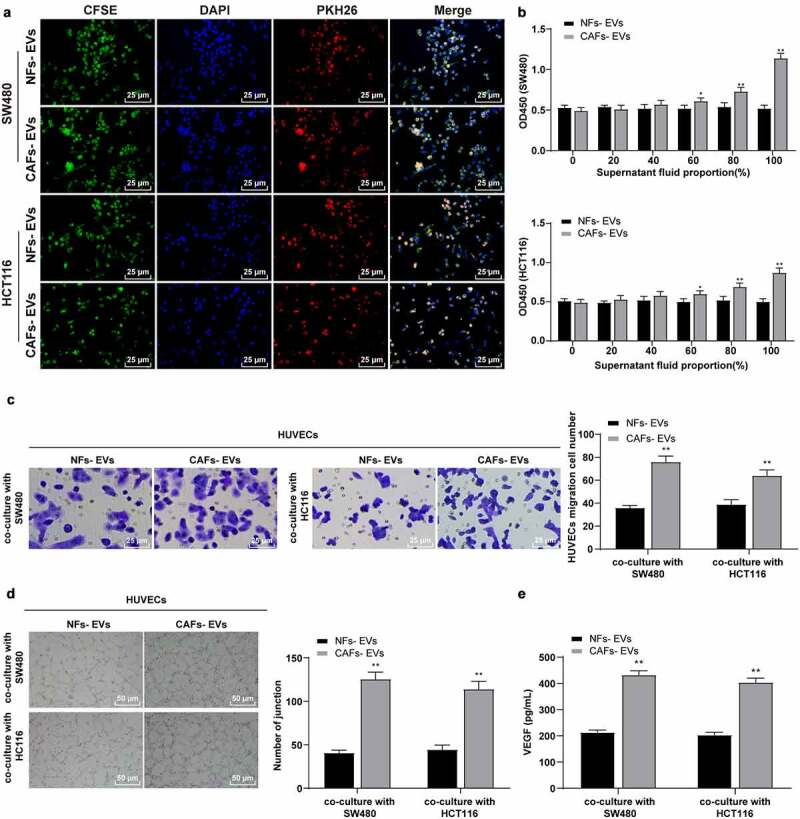
EVs promoted the proliferation, migration, and angiogenesis of HUVECs induced by colon cancer cells. COAD cell lines SW480 and HCT116 were cultured and co-incubated with CAFs-EVs or NFs-EVs for 24 h. (a) The uptake of EVs was observed using immunofluorescence. Then, SW480 and HCT116 cells in different EVs treatment groups were co-cultured with HUVECs. (b) The proliferation of HUVECs was measured using CCK-8; (c) The migration was detected using Transwell assay; (d) The number of connections was measured to quantify the tube formation of HUVECs; (e) The VEGF content in HUVECs supernatant was detected using ELISA. Three cell tests were performed and the data were expressed as mean ± standard deviation. Independent t test was used for comparison among multiple groups. *P < .05, **P < .01.
The revised figure 2 in the attachment shall prevail. Thank you.
CAFs-EVs carried miR-135b-5p into COAD cells
An existing study has identified a close association between miR-135b-5p and angiogenesis. The upregulation of miR-135b-5p can improve the angiogenesis ability of hUC-MSC-EVs15 and promote endothelial cell proliferation and angiogenesis in diabetic retinopathy mice.14 Previously, a high expression pattern of miR-135b-5p has been determined in colorectal cancer.16 An analysis of the ECORI Pan-Cer database showed that miR-135b-5p was highly expressed in COAD (Figure 3a). We further speculated a definitive role of CAFs-EVs in the angiogenesis of COAD by transporting miR-135b-5p. After subsequent RNase treatment, no significant difference was evident in the expression pattern of miR-135b-5p in CAFs-EVs, while the miR-135b-5p expression pattern was downregulated significantly after the addition of SDS during RNase treatment (all P < .01) (Figure 3b), thereby indicating that miR-135b-5p was encapsulated in CAFs-EVs.
Figure 3.
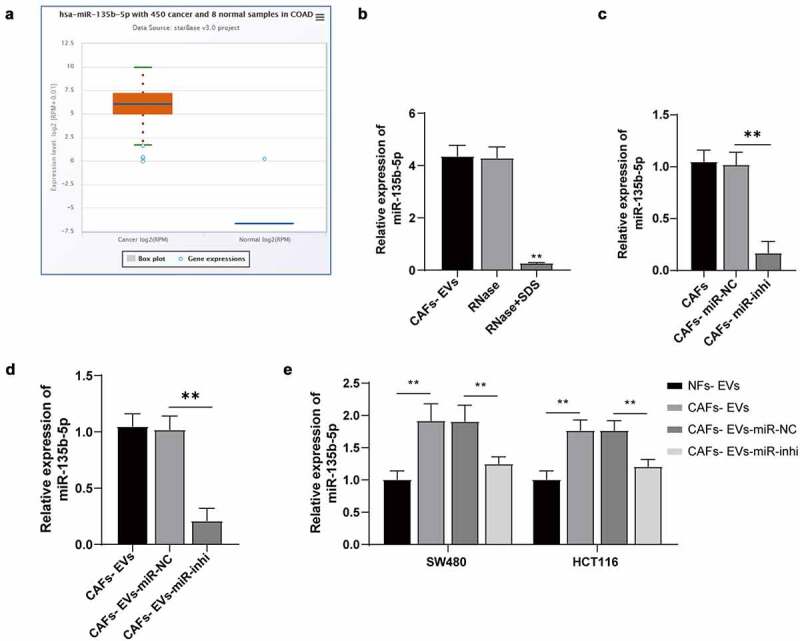
CAFs-EVs carried miR-135b-5p into COAD cells. (a) The miR-135b-5p expression pattern in COAD was analyzed using the ECORI Pan-Cer database. The CAFs and EVs were treated differently, and (b–d) The expression pattern of miR-135b-5p was detected by RT-qPCR. SW480 and HCT116 cells were treated with EVs of different treatment groups. (e) miR-135b-5p expression pattern in COAD cells was detected by RT-qPCR. Three cell tests were performed and the data were expressed as mean ± standard deviation. One-way ANOVA was used for comparison among groups. Tukey’s multiple comparisons test was used for the post hoc test. **P < .01.
Additionally, CAFs were transfected with the miR-135b-5p inhibitor to inhibit the miR-135b-5p expression pattern and facilitate isolation of EVs, and the miR-135b-5p expression pattern in CAFs and CAFs-EVs was both significantly downregulated (all P < .01) (Figure 3c–d). After treatment of SW480 and HCT116 cells with CAFs-EVs-inhi, the expression pattern of miR-135b-5p in COAD cells was significantly downregulated compared with the expression pattern in the CAFs+EVs-miR-NC group (all P < .01) (Figure 3e). The aforementioned results indicated that CAFs-EVs delivered miR-135b-5p into COAD cells.
Inhibition of miR-135b-5p in EVs partially reversed the promotive effect of EVs on angiogenesis induced by COAD cells
To further explore whether CAFs-EVs played a role in the angiogenesis of COAD through miR-135b-5p, the SW480/HCT116 cells in different treatment groups were co-cultured with HUVECs, with detection of the proliferation, migration, and angiogenesis of HUVECs. In comparison with the EVs treatment, inhibition of miR-135b-5p in EVs significantly reduced the proliferation and migration of HUVECs (P < .01) (Figure 4a–b) and the angiogenesis ability (P < .01) (Figure 4c–d). The aforementioned results indicated that inhibition of miR-135b-5p in EVs partially annulled the effect of EVs on enhancing the proliferation, migration, and angiogenesis of HUVECs mediated by COAD cells, suggesting that EVs exerted the effect by delivering miR-135b-5p into COAD cells.
Figure 4.
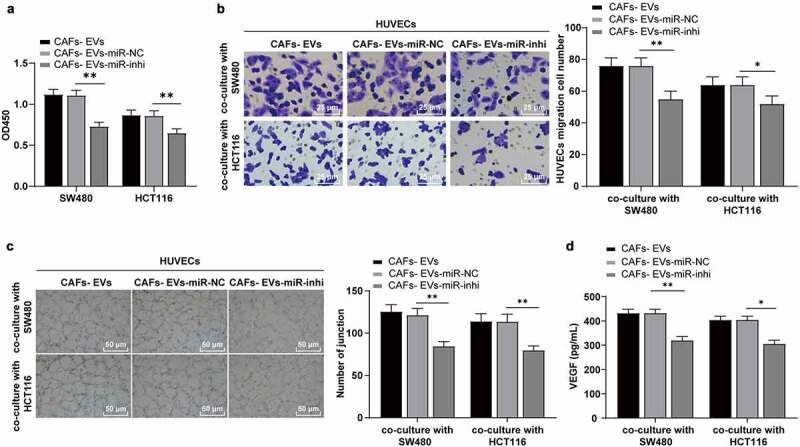
The inhibition of miR-135b-5p in EVs partially reversed the effect of EVs on promoting angiogenesis induced by COAD cells. SW480/HCT116 cells treated with EVs of different treatment groups were co-cultured with HUVECs. (a) The proliferation of HUVECs was detected using CCK-8; (b) The migration was detected using Transwell; (c) The number of connections was measured to quantify the tube formation of HUVECs on the substrate; (d) The VEGF content in HUVECs supernatant was detected using ELISA. Three cell tests were performed and the data were expressed as mean ± standard deviation. One-way ANOVA was used for comparison among multiple groups. Tukey’s multiple comparisons test was used for the post hoc test. *P < .05, **P < .01.
miR-135b-5p targeted FOXO1
To further explore the regulatory mechanism of miR-135b-5p in EVs promoting COAD cell-mediated angiogenesis, the downstream target genes of miR-135b-5p and their intersections were predicted using a combination of StarBase, RNAInter, Targetscan, and miRDB database (Figure 5a). The co-expression relationships of genes were analyzed to further screen target genes using the Coexpedia database and 3 target genes ELK3, MEF2C, and FOXO1 with the highest score (score = 78.213, 71.840, 52.113) were screened according to the co-expression score of the website (Figure 5b). An existing study identified FOXO1 as a transcription factor and a vital regulator of angiogenesis.17 Moreover, the analysis of the ECORI Pan-Cer database demonstrated notable downregulation of FOXO1 in COAD (Figure 5c), and the expression pattern of FOXO1 was negatively correlated with miR-135b-5p (Figure 5d). Starbase prediction identified a targeted binding relationship between miR-135b-5p and FOXO1 3ʹUTR (Figure 5e). The targeted binding relationship between miR-135b-5p and FOXO1 was verified by means of dual-luciferase assay (P < .01) (Figure 5f). Furthermore, the FOXO1 mRNA expression pattern in the co-culture system of SW480/HCT116 and HUVECs was detected. After CAFs-EVs treatment, the expression pattern of FOXO1 mRNA in the co-culture system was notably downregulated, while the FOXO1 mRNA expression pattern was significantly upregulated after inhibition of miR-135b-5p in CAFs-EVs (P < .01) (Figure 5g), hereby eliciting a negative regulatory relationship between miR-135b-5p and FOXO1.
Figure 5.
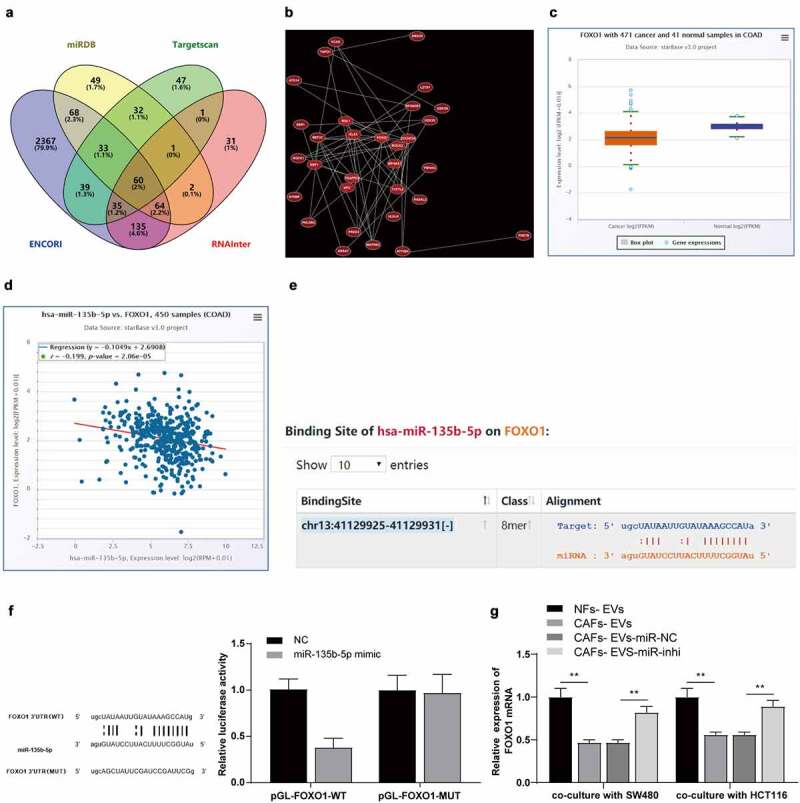
miR-135b-5p targeted FOXO1. (a) Venn map of StarBase, RNAInter, Targetscan, and miRDB database were used to predict the connections of miR-135b-5p downstream target genes; (b) Co-expression network of candidate genes; (c–d) ECORI Pan-Cer database was used to analyze the expression pattern of FOXO1 in COAD and its correlation with miR-135b-5p; (e) StarBase was used to predict and analyze the targeted binding relationship between miR-135b-5p and FOXO1; (f) The targeted binding was verified by dual-luciferase reporter assay; (g) The expression of FOXO1 mRNA in SW480/HCT116 co-cultured with HUVECs was detected by RT-qPCR. Three cell tests were performed and the data were expressed as mean ± standard deviation. Independent t test was used for comparison of the data in F and one-way ANOVA was used for comparison of the data in G. Tukey’s multiple comparisons test was used for the post hoc test. **P < .01.
The revised figure 5 in the attachment shall prevail. Thank you.
Overexpression of FOXO1 reversed the effect of EVs on promoting angiogenesis of COAD cells
Furthermore, FOXO1 was overexpressed after treatment of SW480/HCT116 cells with CAFs-EVs. WB demonstrated a markedly elevated expression pattern of FOXO1 in the co-culture system (P < .01) (Figure 6a). Subsequently, the proliferation, migration, and angiogenesis abilities of HUVECs were detected. Our results denoted that the overexpression of FOXO1 annulled the effects of EVs on promoting the proliferation, migration, and angiogenesis of HUVECs (P < .01) (Figure 6b–e). The preceding results indicated that CAFs-EVs downregulated FOXO1 by carrying miR-135b-5p into COAD cells, thus effectively promoting HUVECs proliferation, migration, and angiogenesis.
Figure 6.
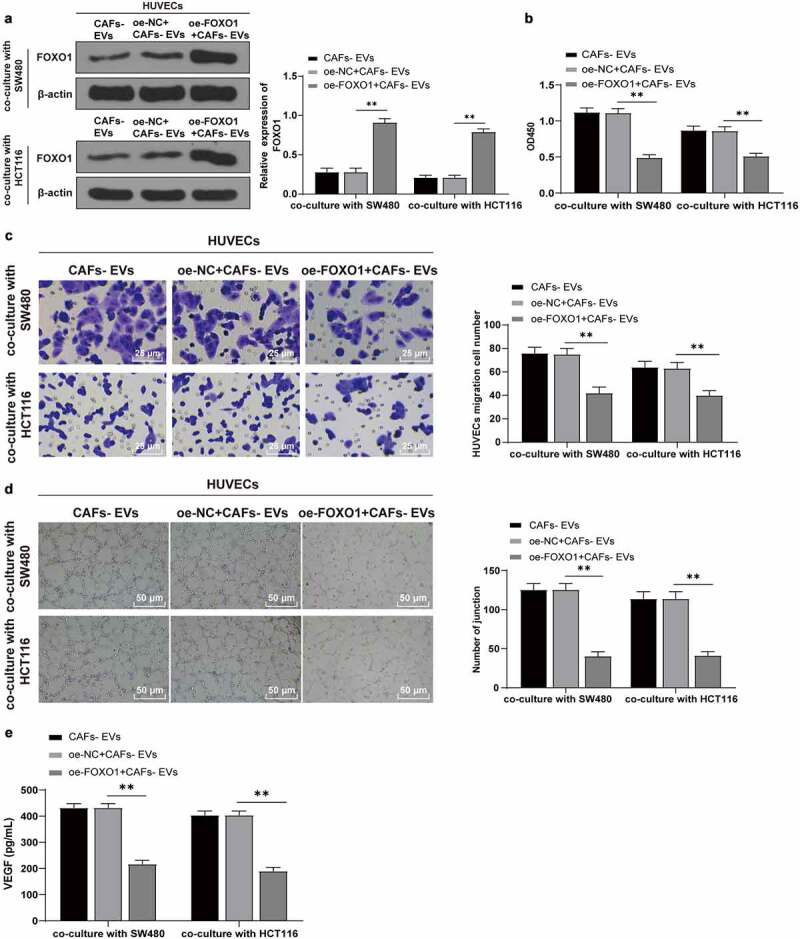
Overexpression of FOXO1 reversed the effect of EVs on promoting angiogenesis. The FOXO1 was overexpressed in CAFs-EV-treated SW480/HCT116 cells. (a) The expression pattern of FOXO1 was detected using WB; (b) The proliferation of HUVECs was detected using CCK-8; (c) The migration was detected using Transwell; (d) The number of connections was measured to quantify the tube formation of HUVECs; (e) The VEGF content in HUVECs supernatant was detected using ELISA. Three cell tests were performed and the data were expressed as mean ± standard deviation. One-way ANOVA was used for comparison among groups. Tukey’s multiple comparisons test was used for the post hoc test. **P < .01.
CAFs-EVs promoted angiogenesis of xenograft tumors by mediating miR-135b-5p
COAD cells SW480 were selected to establish a mouse model of COAD. The mice were injected with EVs under different treatment groups via the tail vein. The tumor growth was observed and the tumor size was measured every 7 days. The inhibition of miR-135b-5p in CAFs-EVs suppressed tumor growth rate, volume, and weight (P < .01) (Figure 7a–c). In order to detect the effect of CAFs-EVs on tumor proliferation and angiogenesis, the tissue sections were stained with Ki67, VEGF, and CD34 antibodies. Our results illustrated that compared with the COAD+CAFs-EVs-NC group, the Ki67, VEGF, and CD3 positive cells were decreased after CAFs-EVs-miR-inhi treatment (Figure 7d–e). ELISA further verified the ability of CAFs-EV-miR-inhi treatment to significantly decrease the content of VEGF in the tumor tissues and repressed angiogenesis (P < .01) (Figure 7f). The tumor samples were detected by RT-qPCR and WB and compared with the COAD + CAFs-EVs-miR-NC group, the expression pattern of miR-135b-5p was significantly downregulated, while the expression pattern of FOXO1 was significantly upregulated after CAFs-EVs-miR-inhi treatment (all P < .01) (Figure 7g–h).
Figure 7.
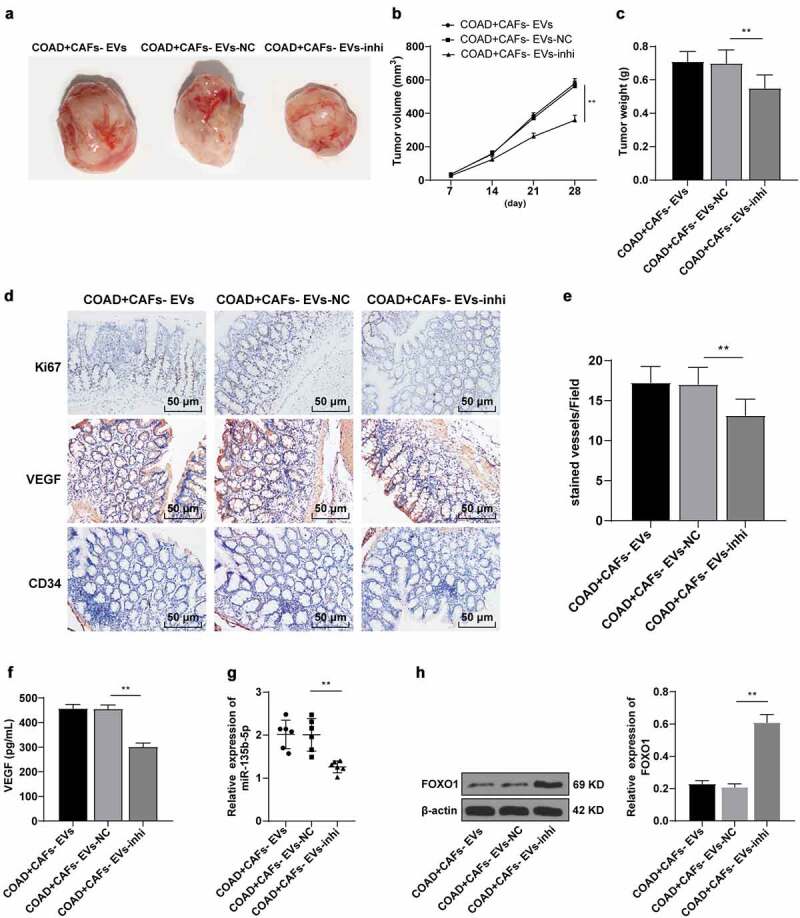
CAFs-EVs promoted angiogenesis of xenograft tumors in vivo. The nude mouse model of COAD was established by transplanting SW480 cells into the skin of nude mice. The mice were injected with CAFs-EVs of different treatment groups through the tail vein. (a–c) The growth rate, volume, and weight of the tumor were observed and measured. The tumor tissues were collected. (d) The expression patterns of Ki67, VEGF, and CD34 were detected using immunohistochemistry; (e) Microvessel density (MVD) was reflected by the number of CD34 microvessels; (f) The VEGF content in HUVECs supernatant was detected using ELISA; (g) The expression of miR-135b-5p was detected by RT-qPCR; (h) The expression pattern of FOXO1 was detected by WB. N = 6; the data were expressed as mean ± standard deviation. One-way ANOVA was used for comparison among multiple groups. Tukey’s multiple comparisons test was used for the post hoc test. **P < .01.
Discussion
COAD is the most prevalent type of tumor worldwide and is regarded among the most explicit causes of cancer associated mortality, along with breast, lung, and prostate cancers.20 Limited evidence has identified the involvement of EVs in the development of COAD.21 The current study revealed that CAFs-EVs had promoted the angiogenesis of COAD cells via regulation of miR-135b-5p/FOXO1.
CAFs-EVs potentiate cell malignant biological behaviors in cancers.8 The uptake assay validated the internalization of both CAFs-EVs and NFs-EVs by SW480 and HCT116 cells. In order to determine the effect of CAFs-EVs on COAD, the SW480 and HCT116 cell supernatants treated with CAFs-EVs or NFs-EVs were co-cultured with HUVECs. Our experimental results demonstrated that CAFs-EVs had effectively promoted COAD cell-mediated HUVEC proliferation, migration, and angiogenesis, while NFs-EVs had no significant effects on HUVECs. Research has identified VEGF as a vital factor for angiogenesis.22 Our results demonstrated significantly elevated VEGF content in HUVECs supernatant co-cultured with CAFs-EVs-treated COAD cells. Additionally, an existing study had determined the ability of EVs to manipulate fundamental processes trivial for tumor progression including cell proliferation, angiogenesis, invasion, and migration.23 Consistently, EVs derived from cancer cells are essential for inducing malignant behaviors in COAD cells.24 Briefly, our findings elicited that CAFs-EVs induced COAD cell-mediated HUVECs malignant biological behaviors and angiogenesis.
The circulating EVs can transport miR byproducts to the surrounding cells, thus facilitating intercellular communication between different cells for a significant physiological effect.25 An existing study determined a close association between miR-135b-5p and angiogenesis and miR-135b-5p upregulation improves the angiogenic capacity of hUC-MSC-derived EVs.15 Our results showed a notably upregulated miR-135b-5p expression in COAD, which was in consistency with previously reported findings.16 In light of the aforementioned literature, we speculated that CAFs-EVs might manipulate effects on COAD by transporting miR-135b-5p. Our results revealed that miR-135b-5p was encapsulated in CAFs-EVs, which led to a downregulated miR-135b-5p expression in COAD cells after CAFs-EVs-inhi treatment. Consistently, these results demonstrated complete transport of miR-135b-5p by CAFs-EVs into COAD cells. To further investigate the function of CAFs-EVs in COAD through miR-135b-5p, the CAFs-EVs-treated SW480/HCT116 cells were co-cultured with HUVECs. Our findings demonstrated partial annulment of the effects of EVs in inhibiting the malignant behaviors of HUVECs after inhibition of the miR-135b-5p expression in CAFs-EVs. Consistently, miR-135b overexpression can facilitate the progression and transformation of COAD, while inhibition of miR-135b can weaken tumor growth by manipulation of the regulatory genes implicated in the malignant behaviors in COAD.26 Altogether, our findings elicited that CAFs-EVs inhibited the malignant behaviors of HUVECs by transporting miR-135b-5p into COAD cells. COAD cells SW480 were used to establish a mouse model of COAD, followed by injection with EVs in different groups. miR-135b-5p inhibition in CAFs-EVs suppressed tumor growth rate, volume, and weight, decreased the Ki67, VEGF, and CD3 positive cells, and significantly reduced the content of VEGF and repressed angiogenesis. These results suggest that CAFs-EVs promote COAD proliferation and angiogenesis by transporting miR-135b-5p.
To study the downstream mechanism of miR-135b-5p, a combination of StarBase, RNAInter, Targetscan, and miRDB database were adopted to predict the downstream target genes of miR-135b-5p, after which FOXO1 was identified. Fundamentally, FOXO1 is a forkhead transcription factor that functions as an essential regulator of angiogenesis.17 ECORI Pan-Cer database elicited that FOXO1 was downregulated in COAD. Our findings validated a negatively correlated relationship between miR-135b-5p and FOXO1 along with the targeted binding relationship. Consistently, FOXO1 has been identified as the potential downstream target of miR-135b-5p in colorectal cancer.27 Our results elicited notable downregulation of FOXO1 in the co-culture system after CAFs-EV treatment, however it was markedly upregulated after inhibition of miR-135b-5p in CAFs-EVs. Moreover, we overexpressed FOXO1 in CAFs-EVs-treated SW480/HCT116 cells. Our results were illustrative of conflicting CAFs-EVs effect on the malignant behaviors of HUVECs cells after overexpression of FOXO1. Relative to the cells with knockdown of FOXO1, the apoptosis rate is higher in the presence of FOXO1, moreover, the FOXO1 degradation induces COAD cell proliferation.28 In conclusion, CAFs-EVs targeted FOXO1 and promoted HUVEC malignant behaviors by delivering miR-135b-5p into COAD cells.
In summary, the findings of the current study supported that CAFs-EVs-shuttled miR-135b-5p targeted FOXO1, thereby inducing the malignant behaviors and angiogenesis of COAD cells-mediated HUVECs cells. However, the direct effect of EVs on HUVECs, and other miRNAs that EVs could potentially regulate, other target genes under potential regulation of miR-135b-5p, and the downstream pathways that FOXO1 might regulate were not investigated. Moreover, the effect of EVs has not been studied at the clinical level. Future studies are warranted to identify the upstream lncRNAs in EVs that regulate miR-135b-5p or the role and mechanism of other EV-carried lncRNAs and miRNAs in COAD, or other downstream target genes and relative pathways that miR-135b-5p may potentially regulate.
Funding Statement
This work was supported by Ningbo Clinical Research Center for Digestive System Tumors (Ningbo Clinical Reserch Center for Digestive System Tumors 2019A21003), the Natural Public Welfare Fund of Zhejiang Province (LGC20H160002), the Medical and Health Science and Technology Foundation of Zhejiang Province (2019KY595, 2018KY690, 2018KY699, 2017KY593, 2017KY594), the Natural Science Foundation of Ningbo(2018A610368, 2017A610145, 2017A610158, 2016A610135).
Availability of data and materials
All the data generated or analyzed during this study are included in this published article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
Conceptualization: XYD and YYX; validation, research, resources, data reviewing, and writing: XYD, YYX, MJD; review and editing: YYX and MJD. All authors read and approved the final manuscript.
References
- 1.Karpisheh V, Nikkhoo A, Hojjat-Farsangi M, Namdar A, Azizi G, Ghalamfarsa G, Sabz G, Yousefi M, Yousefi B, Jadidi-Niaragh F, et al. Prostaglandin E2 as a potent therapeutic target for treatment of colon cancer. Prostaglandins Other Lipid Mediat. 2019;144:106338. DOI: 10.1016/j.prostaglandins.2019.106338. [DOI] [PubMed] [Google Scholar]
- 2.Skh L, Martin A.. Mismatch repair and colon cancer: mechanisms and therapies explored. Trends Mol Med. 2016;22(4):274–289. DOI: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22:12. DOI: 10.1186/s11658-017-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Bhatt LK, Johnston TP, Prabhavalkar KS. Colon cancer stem cells: potential target for the treatment of colorectal cancer. Cancer Biol Ther. 2019;20(8):1068–1082. DOI: 10.1080/15384047.2019.1599660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai YP, Shang K, Chen H, Ding F, Wang Z, Liang C, Xu, Y, Sun , M, Y, Y. FGF-1/-3/FGFR4 signaling in cancer-associated fibroblasts promotes tumor progression in colon cancer through Erk and MMP-7. Cancer Sci. 2015;106:1278–1287. DOI: 10.1111/cas.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unterleuthner D, Neuhold P, Schwarz K, Janker L, Neuditschko B, Nivarthi H, Crncec I, Kramer N, Unger C, Hengstschläger M, et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis. 2020;23(2):159–177. DOI: 10.1007/s10456-019-09688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ando N, Hara M, Shiga K, Yanagita T, Takasu K, Nakai N, Maeda, Y, Hirokawa, T, Takahashi, H, Ishiguro, H, et al. Eicosapentaenoic acid suppresses angiogenesis via reducing secretion of IL6 and VEGF from colon cancerassociated fibroblasts. Oncol Rep. 2019;42:339–349. DOI: 10.3892/or.2019.7141. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, Li B, Yang J, Shen J, Ji J, Miao M. Fibroblastderived exosomal microRNA369 potentiates migration and invasion of lung squamous cell carcinoma cells via NF1mediated MAPK signaling pathway. Int J Mol Med. 2020;46:595–608. DOI: 10.3892/ijmm.2020.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. DOI: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 10.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang Z-Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20. DOI: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. DOI: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67(3):940–954. DOI: 10.1002/hep.29586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi X, Teng M, Zhang L, Xia L, Chen J, Cui Z. Retracted: microRNA-204-3p represses colon cancer cells proliferation, migration, and invasion by targeting HMGA2. J Cell Physiol. 2020;235(2):1330–1338. DOI: 10.1002/jcp.29050. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Xu H, Zhao H, Sui D. MicroRNA-135b-5p promotes endothelial cell proliferation and angiogenesis in diabetic retinopathy mice by inhibiting Von Hipp-el-Lindau and elevating hypoxia inducible factor alpha expression. J Drug Target. 2021;29(3):300–309. DOI: 10.1080/1061186X.2020.1833017. [DOI] [PubMed] [Google Scholar]
- 15.Yang K, Li D, Wang M, Xu Z, Chen X, Liu Q, Sun W, Li J, Gong Y, Liu D, et al. Exposure to blue light stimulates the proangiogenic capability of exosomes derived from human umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2019;10(1):358. DOI: 10.1186/s13287-019-1472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proenca MA, Biselli JM, Succi M, Severino FE, Berardinelli GN, Caetano A, Reis RM, Hughes DJ, Silva AE. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. 2018;24(47):5351–5365. DOI: 10.3748/wjg.v24.i47.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren B. FoxO1 transcriptional activities in VEGF expression and beyond: a key regulator in functional angiogenesis? J Pathol. 2018;245(3):255–257. DOI: 10.1002/path.5088. [DOI] [PubMed] [Google Scholar]
- 18.Qin Y, Li L, Wang F, Zhou X, Liu Y, Yin Y, Qi X. Knockdown of Mir-135b sensitizes colorectal cancer cells to oxaliplatin-induced apoptosis through increase of FOXO1. Cell Physiol Biochem. 2018;48(4):1628–1637. DOI: 10.1159/000492284. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Wang Y, Yan Z, Sun Y, Su C. Colorectal cancer cell-derived exosomes promote proliferation and decrease apoptosis by activating the ERK pathway. Int J Clin Exp Pathol. 2019;12(7):2485–2495. [PMC free article] [PubMed] [Google Scholar]
- 20.Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, et al. Colon cancer. Crit Rev Oncol Hematol. 2010;74(2):106–133. DOI: 10.1016/j.critrevonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Zhong ME, Chen Y, Xiao Y, Xu L, Zhang G, Lu J, Qiu H, Ge W, Wu B. Serum extracellular vesicles contain SPARC and LRG1 as biomarkers of colon cancer and differ by tumour primary location. EBioMedicine. 2019;50:211–223. DOI: 10.1016/j.ebiom.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176(6):1248–1264. DOI: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan R, Maresh G, Zhang X, Salomon C, Hooper J, Margolin D, Li L. The Emerging Roles of Extracellular Vesicles As Communication Vehicles within the Tumor Microenvironment and Beyond. Front Endocrinol (Lausanne). 2017;8:194. DOI: 10.3389/fendo.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul D, Roy A, Nandy A, Datta B, Borar P, Pal SK, Senapati, D, Rakshit, T. Identification of biomarker hyaluronan on colon cancer extracellular vesicles using correlative AFM and spectroscopy. J Phys Chem Lett. 2020;11(14):5569–5576. DOI: 10.1021/acs.jpclett.0c01018. [DOI] [PubMed] [Google Scholar]
- 25.Maciel E, Mansuy IM. Extracellular vesicles and their miRNA Cargo: a means of communication between soma and germline in the mammalian reproductive system. Chimia (Aarau). 2019;73(5):356–361. DOI: 10.2533/chimia.2019.356. [DOI] [PubMed] [Google Scholar]
- 26.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart J, Ueno L, Grivennikov S, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. DOI: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12(6):5259–5279. DOI: 10.18632/aging.102945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chae YC, Kim JY, Park JW, Kim KB, Oh H, Lee KH, Seo, SB. FOXO1 degradation via G9a-mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019;47(4):1692–1705. DOI: 10.1093/nar/gky1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated or analyzed during this study are included in this published article.


