Figure 2.
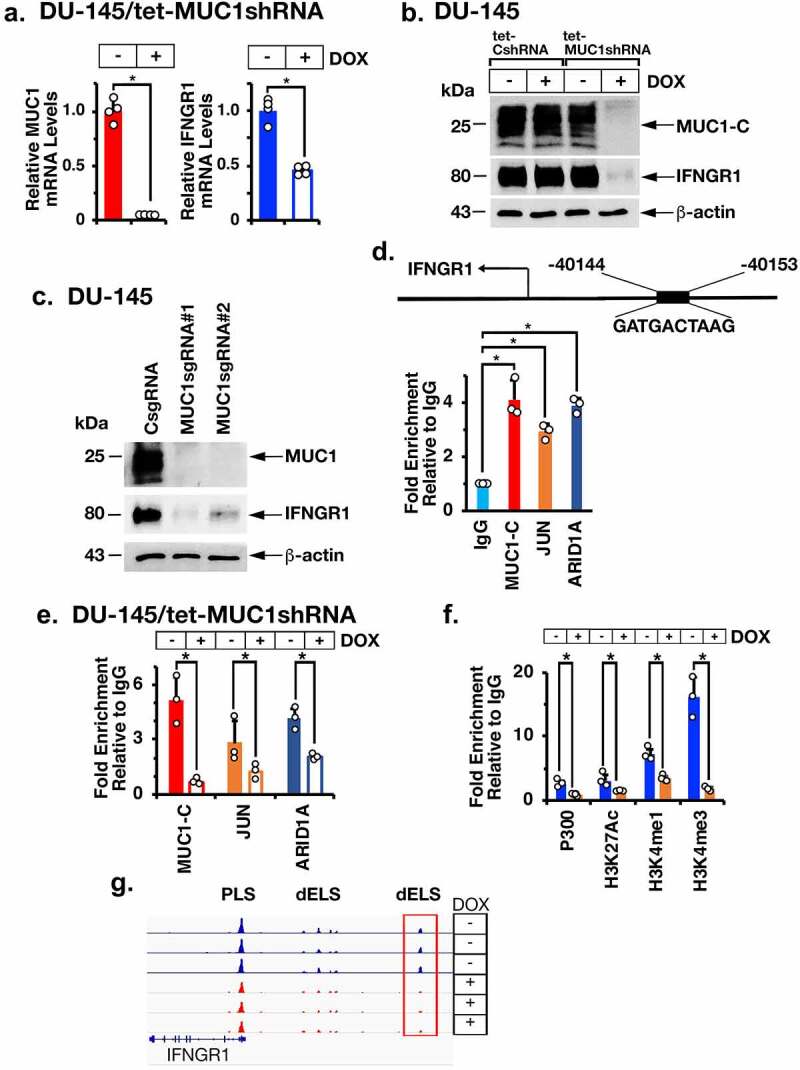
The MUC1-C→ARID1A/BAF pathway is necessary for activation of IFNGR1 expression. (a). DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days were analyzed for MUC1-C and IFNGR1 mRNA levels by qRT-PCR using primers listed in Supplemental Table S1. The results (mean ± SD of 4 determinations) are expressed as relative mRNA levels compared to that obtained for vehicle-treated cells (assigned a value of 1). (b). Lysates from DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA treated with vehicle or DOX for 7 days were immunoblotted with antibodies against the indicated proteins. (c). Lysates from DU-145 expressing a CsgRNA, MUC1sgRNA#1 or MUC1sgRNA#2 were immunoblotted with antibodies against the indicated proteins. (d). Schema of the IFNGR1 with highlighting of a JUN/AP-1 binding site in the dELS. Soluble chromatin from DU-145 cells was precipitated with a control IgG, anti-MUC1-C, anti-JUN and anti-ARID1A. (e). Soluble chromatin from DU-145/tet-MUC1shRNA cells treated with vehicle or DOX was precipitated with a control IgG, anti-MUC1-C, anti-JUN and anti-ARID1A. (f). Soluble chromatin from DU-145/tet-MUC1shRNA cells treated with vehicle or DOX was precipitated with a control IgG, anti-EP300, anti-H3K27ac, anti-H3K27me1 and anti-H3K4me3. The DNA samples were amplified by qPCR with primers for the IFNGR1 dELS region. The results (mean ± SD of 3 determinations) are expressed as fold enrichment relative to that obtained with the IgG control (assigned a value of 1). (g and h). Genome browser snapshots of ATAC-seq data from the IFNGR1 dELS in DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days (g). Chromatin was analyzed for accessibility by nuclease digestion (h). The results (mean ± SD of 3 determinations) are expressed as % untreated chromatin.
