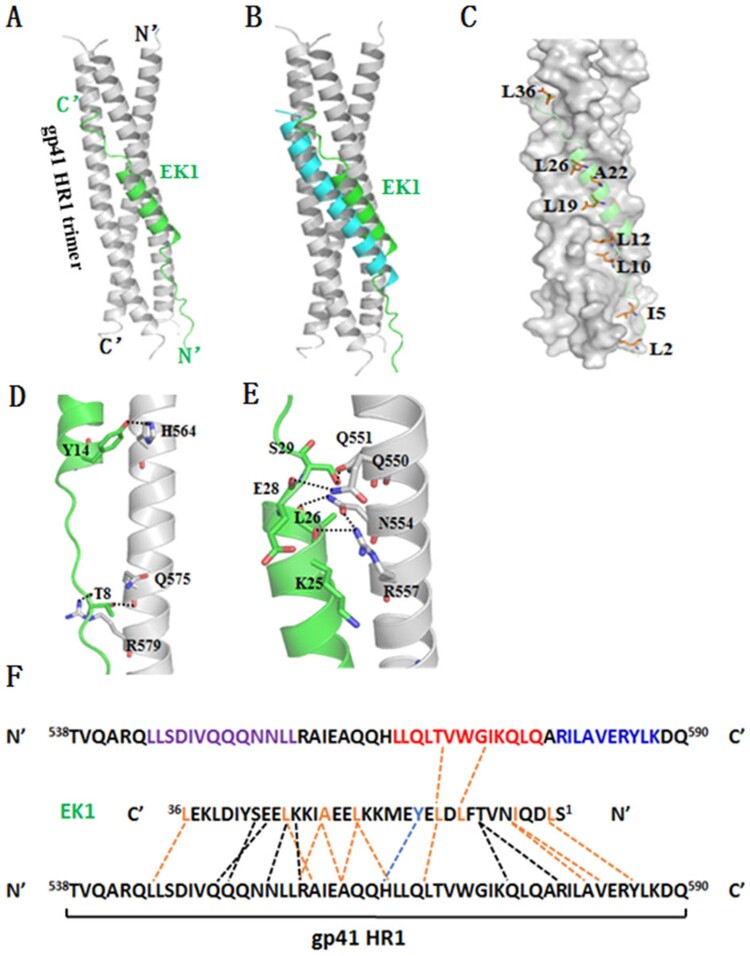
Figure 5.
Binding model of EK1 peptide with the HR1 of HIV-1 gp41 by molecular docking. The residues involving hydrogen bonds and hydrophobic interaction are shown as stick models with labels. Hydrogen bonds and PI-bond are indicated in dashed lines. (A) A ribbon model of EK1/HR1 structure, in which the HR1 trimer is coloured in grey and EK1 is in green. (B) Superimposing of EK1 with the gp41 HR2 peptide C34 (in cyan). (C) A group of hydrophobic residues of EK1 make extensive contacts with the gp41 HR1 surface critically determining the inhibitor binding. (D) At the extended N terminal of EK1, T8 donates a hydrogen to the gp41 Gln-575 while it accepts a hydrogen from the gp41 Arg-579. At the beginning of EK1 helix portion, a PI-bond is formed between Y14 of EK1 and His-564 of gp41. (E) Around the end of the EK1 helix, a hydrogen-bond network encompasses four pairs of interactions: the long side chain of Arg-557 donates a hydrogen bond to the O_atom of K25, the side chain of Asn-554 donates a hydrogen bond to the O_atom of L26, the side chain of Gln-550 donates a hydrogen bond to the O_atom of E28, and the side chain of Gln-551 accepts a hydrogen bond from S29. (F) Sequence illustration of EK1 binding modelled by molecular docking. A single EK1 peptide interacting with two NHR helices is shown in a sequence map. The dashed black lines indicate the interhelical hydrogen bonds, the dashed blue line indicates a pi-bond, and the dashed orange lines indicate hydrophobic interactions. The sequences mediating T20 resistance, pocket-1 site, and pocket-2 site are marked in purple, red, and blue, respectively