SUMMARY
Cells from across the eukaryotic tree use actin po ymer networks for a wide variety of functions including endocytosis, cytokinesis, and cell migration. Despite this functional conservation, the actin cytoskeleton has undergone significant diversification, highlighted by the differences in the actin networks of mammalian cells and yeast. Chytrid fungi diverged before the emergence of the Dikarya (multicellular fungi and yeast), and therefore provide a unique opportunity to study actin cytoskeletal evolution. Chytrids have two life stages: zoospore cells that can swim with a flagellum, and sessile sporangial cells that, like multicellular fungi, are encased in a chitinous cell wall. Here we show that zoospores of the amphibian-killing chytrid Batrachochytrium dendrobatidis (Bd) build dynamic actin structures resembling those of animal cells, including an actin cortex, pseudopods, and filopodia-like spikes. In contrast, Bd sporangia assemble perinuclear actin shells and actin patches similar to those of yeast. The use of specific small molecule inhibitors indicate that nearly all of Bd’s actin structures are dynamic and use distinct nucleators: while pseudopods and actin patches are Arp2/3-dependent, the actin cortex appears formin-dependent, and actin spikes require both nucleators. Our analysis of multiple chytrid genomes reveal actin regulators and myosin motors found in animals but not dikaryotic fungi, as well as fungal-specific components. The presence of animal- and yeast-like actin cytoskeletal components in the genome combined with the intermediate actin phenotypes in Bd suggests that the simplicity of the yeast cytoskeleton may be due to evolutionary loss.
eTOC Blurb
Prostak et al. use early-diverging chytrid fungi to investigate actin cytoskeletal evolution. They find that chytrids have animal-like actin cytoskeleton genes lost by multicellular fungi and that the frog-killing chytrid B. dendrobatidis has actin networks similar in struc ure and regulation to those of both animal cells and multicellular fungi.
INTRODUCTION
Actin participates in nearly every essential eukaryotic cell function, including endocytosis, intracellular trafficking, cell migration, and cytokinesis in many species. Eukaryotic cells employ a sophisticated network of actin regulatory proteins to spatially and temporally control these diverse functions1. How these complex actin regulatory networks evolved and diversified remain key questions in both evolutionary and cell biology. Here we use chytrids—early-diverging fungi that still share important features of animal cells lost in yeast and other fungi2—as a system to explore the evolution of the actin cytoskeleton. Using a combination of genomics and fluorescence microscopy, we show that chytrid fungi have an actin cytoskeleton that combines features of animal cells and yeast.
Actin polymerization is largely regulated by controlling the initiation of new actin polymers—a process called “actin nucleation.” Actin is nucleated by two main systems: the Arp2/3 complex or formin family proteins, both of which were likely present in the last common eukaryotic ancestor3. While the Arp2/3 complex primarily builds actin branches along the side of existing actin filaments1, formins assemble unbranched filament networks through processive addition of actin monomers by their Formin Homology 2 (FH2) domains1,4.
Animal cells use the Arp2/3 complex and formins to build a wide variety of dynamic actin structures. These structures include diverse membrane protrusions used for movement, from broad, branched-actin-filled pseudopods and lamellipodia, to finger-like filopodia that are packed with linear actin bundles1,5. Dynamic actin networks and their associated motors also mediate membrane invagination during endocytosis as well as during cell division by constriction of a ring of actin called the “cytokinetic ring”1. Many of these actin structures often assemble in proximity to the “actin cortex,” a 200 nm thick actin shell that lies just below and supports the plasma membrane6,7.
Budding and fission yeast, in contrast to animal cells, each have simplified actin networks that consist of three main structures: actin patches that are sites of endocytosis; actin cables for vesicle trafficking and establishing cell polarity; and the cytokinetic actin ring8. Fission yeast also have a fourth actin structure; the fusion focus that is used to deliver cell wall degrading enzymes during sexual reproduction9. This simplification is echoed by a streamlined actin regulatory system that is missing key actin regulators import nt for human health10, particularly the SCAR/WAVE complex which helps drive cell migration involved in normal development of mouse embryos, and in metastasis and tissue invasion in tumor models11. The wide gap between mammalian and yeast actin biology makes it difficult to know which rules of yeast actin regulation apply to human cell biology. Bridging this gap would allow us to apply our deep understanding of simplified yeast actin networks to human cells.
To help us bridge the gap between the simplified actin networks of yeast and the dizzyingly complex actin networks of human cells, we turned to chytrid fungi. Chytrids play key roles in aquatic and terrestrial habitats12, including the amphibian pathogen Batrachochytrium dendrobatidis (Bd) that is decimating global frog populations13. Also known as zoosporic fungi, the >1000 known species of chytrids are likely paraphyletic, comprising at least two fungal phyla: Chytridiomycota and Blastocladiomycota14. Chytrids diverged from a common fungal ancestor before the diversification of the Dikarya14, the group of fungi that includes multicellular mushrooms and sac fungi, as well as unicellular yeasts that descended from multicellular fungi (Figure 1A). Because Dikarya likely arose from within chytrids, these phylogenetic relationships position chytrids as an evolutionary Rosetta Stone with which we can map the simplified actin features of yeast to those of animals.
Figure 1. The chytrid fungus Batrachochytrium dendrobatidis is an early branching fungus with an archetypal chytrid life cycle and animal-like and fungal-like actin structures.
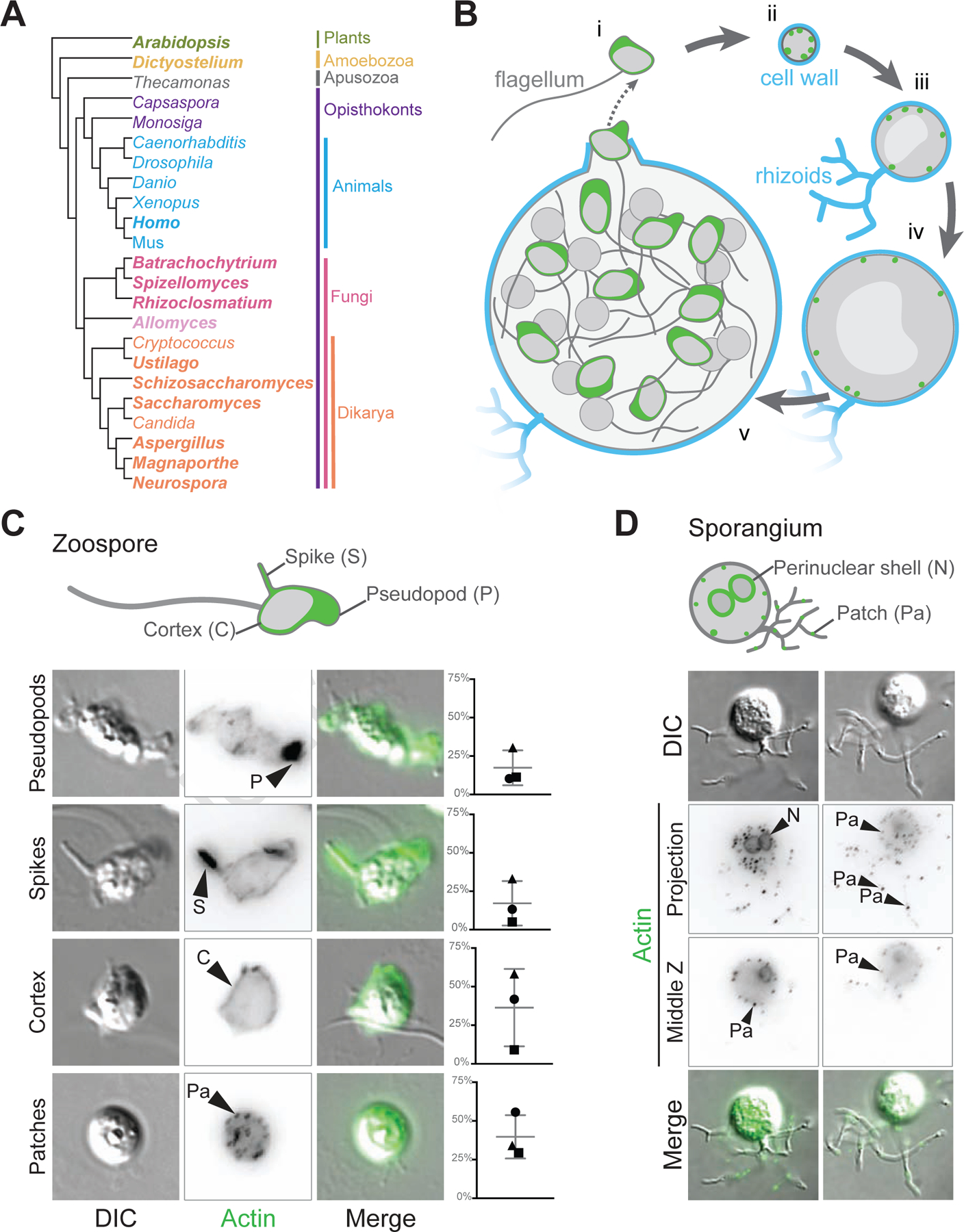
(A) This cladogram shows the relationships between representative genera of major eukaryotic groups. Chytrids are represented by Batrachochytrium, Spizellomyces, Rhizoclosmatium, and Allomyces (magenta, lavender), diverging before the diversification of Dikarya (orange), and are in a sister clade to animals (cyan). Bold type indicates genera used for the majority of the homologous sequence analyses in this paper. (B) In vitro life cycle of Batrachochytrium dendrobatidis (Bd). Bd has a motile stage known as a zoospore (i) with a flagellum made of microtubules, no cell wall, and can crawl using actin (green) based protrusions. Zoospores encyst and build a cell wall (cyan), this stage is referred to as a germling (ii). The germling grows in size, becoming a sporangium. Sporangia develop hyphal-like structures called rhizoids used for nutrient uptake and undergo synchronous rounds of mitosis (iii-iv) before cellularization and release of the next generation of zoos ores (v). This life cycle takes approximately three days in laboratory culture conditions. (C) Representative examples of zoospores (DIC: grey) and the phalloidin stained actin Z-projections of cells at this stage (inverted, black), with an overlay of the two (actin, green). Actin structures in Bd zoospores are: actin-filled pseudopods (P), actin-filled spikes (S), cortical actin (Co), and actin patches (Pa). Graphs on the right indicate the raw percent of cells with the phenotype in 3 independent experiments, shapes here match the shapes for the replicates in Figure 6. (D) Representative examples of Bd sporangia (DIC: grey) and the phalloidin stained actin structures at this stage (inverted, black; both a max intensity z projection and a single slice), with an overlay of the DIC and fluorescence (actin, green). Actin structures in sporangia are: actin patches (Pa), and perinuclear actin shells (N). See also Figure S1.
Bd, like other chytrids, has two developmental stages: motile “zoospores” that lack a cell-wall and swim with a flagellum, and non-motile sporangia that grow and produce new zoospores (Figure 1B)15. The transition from the dispersal stage of the life cycle (the zoospore) to the reproductive phase (the sporangium) coincides with the loss of the flagellum and development of root-like rhizoids that serve to increase the area from which cells can draw nutrients (Figure 1B). We recently showed that zoospores can crawl across surfaces using actin-filled, Arp2/3-dependent pseudopods10, and that cellularization—the process of dividing a single large, multinucleated sporangium into dozens of small, motile zoospores—involves assembly of complex actin networks that visually resemble those that drive cellularization during early Drosophila development2. Here, we identify homologs of key actin regulatory proteins and myosin motors in multiple species of chytrids, including both Chytridiomycota and Blastocladiomycota. We also identify new actin structures in zoospores and sporangia, and test the requirements of Arp2/3 and formin family proteins for their assembly. We find that both the regulatory networks and actin structures of Bd are intermediate in complexity between animals and Dikarya, suggesting that the streamlined actin networks of common model fungi are a result of secondary evolutionary loss of actin network components.
RESULTS
Bd has developmentally distinct actin phenotypes that resemble animal and fungal cells
To determine how Bd may use actin during its life cycle, we stained Bd cells with fluorescent phalloidin that labels polymerized actin10. Staining of Bd zoospores revealed four easily distinguishable actin structures found in various combinations (Figure 1C): (1) pseudopods that are roughly 1–2 µm across; (2) filopodia-like actin “spikes” that have not been previously described in Bd, which we define as thin, actin-filled protrusion <1 µm across and ≥1 µm long; (3) cortical actin localized to more than half of the cell edge (Figure S1); and (4) <1 µm diameter actin puncta. We call these last structures “actin patches” due to their visual similarity to yeast actin patches8 as well as their localization at the cell periphery (Figure 1D). The percent of zoospores with each actin structure is variable from experiment-to-experiment (Figure 1C), likely due to slight differences in developmental staging between biological replicates.
We also stained Bd sporangia—large multinucleated cells that undergo mitosis and expansive cell growth while encased in a cell wall (Figure 1B iii–v). Bd sporangia stained for polymerized actin show two types of actin structures: perinuclear shells, defined by intense F-actin staining around each nucleus, similar to those observed in Spizellomyces punctatus2; and actin patches (Figure 1D). Both structures were present in nearly all observed sporangia.
The actin regulatory network of chytrids resembles that of both animals and Dikarya
To explore how Bd’s animal- and yeast-like actin structures may be built and regulated, we performed BLAST searches for actin and its regulators across five chytrid species: Bd, Batrachochytrium salamandrivorans (Bs), Spizellomyces punctatus (Sp), Rhizoclosmatium globosum (Rg), and Allomyces macrogynus (Am) (Figure 2, Figure S2, Data S1, Data S2), and compared these to homologs from humans, Arabidopsis thaliana, Dictyostelium discoideum, as well as those of dikaryotic fungi: Saccharomyces cerevisiae, Schizosaccharomyces pombe, Schizosaccharomyces japonicus, Candida albicans, Aspergillus nidulans, Magnaporthe oryzae, Neurospora crassa, and Ustilago maydis and summarize these analyses here.
Figure 2. Chytrid actin regulatory protein networks are intermediate to those of animals and Dikarya.
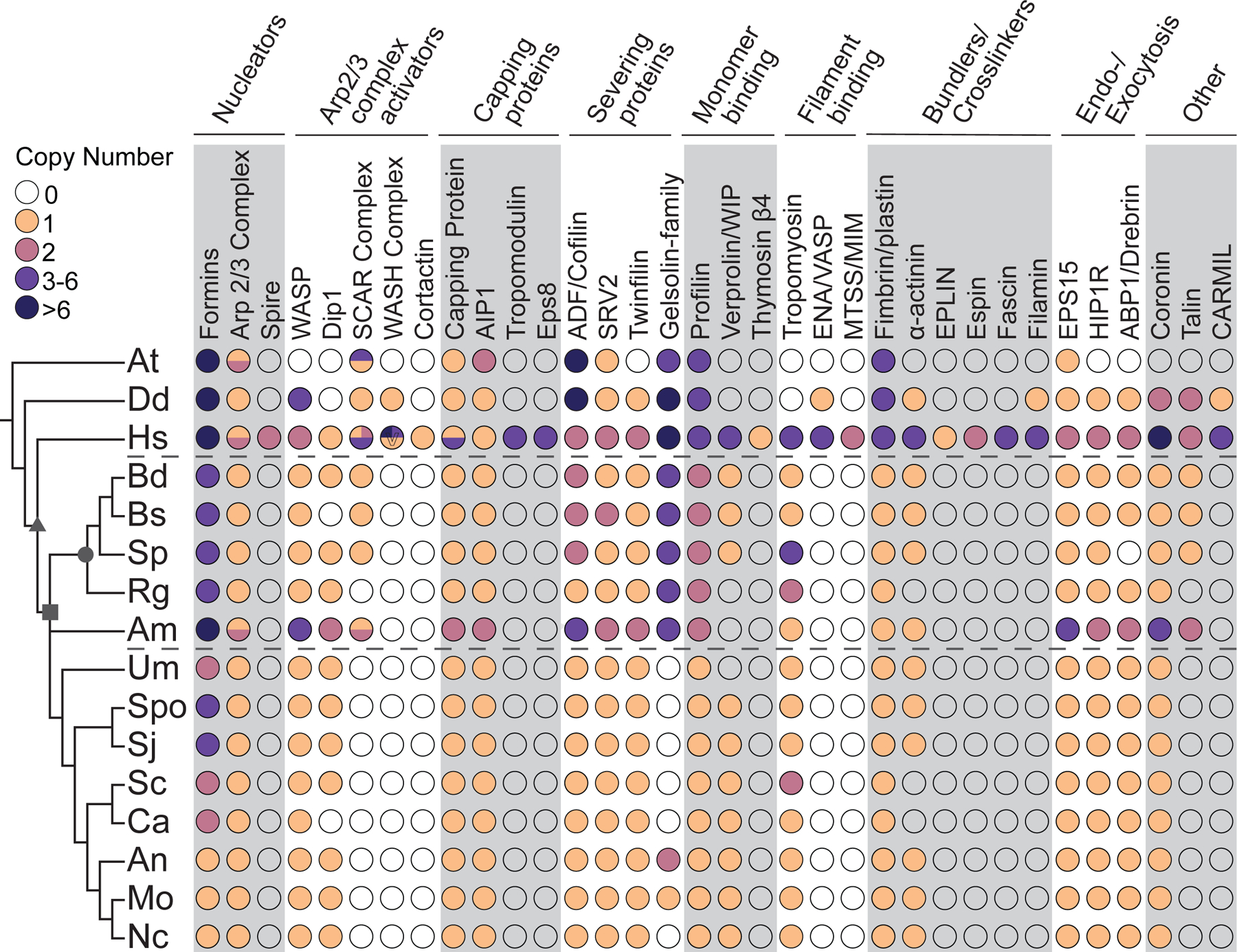
The distribution of actin regulatory proteins across taxa. Color-filled circles indicate the presence of clear homologs found, with the number of homologs for each protein in each species shown in the colors specified in the key. Unfilled circles indicate that no homolog was detected in that species. Circles with multiple colors indicate complexes with different copy numbers for multiple complex members. Circles for Capping Protein represent the copy number for both α and β subunits. Dashed lines mark the chytrids. Symbols on the tree represent: opisthokonts (triangle); fungi (square); chytridiomycota (circle). Symbols in the circles represent: V, copy number of WASH varies individually, as many WASH genes are subtelomeric63. O, Arabidopsis has 5 villin-like genes and an additional gelsolin-domain containing protein, none of which are phylogenetically related to metazoan gelsolin/villin family members64. +, See Data S2 for details and additional potential homologs with caveates. At, Arabidopsis thaliana; Dd, Dictyostelium discoideum; Hs, Homo sapiens; Bd, Batrachochytrium dendrobatidis; Bs, Batrachochytrium salamandrivorans; Sp, Spizellomyces punctatus; Rg, Rhizoclosmatium globosum; Am, Allomyces macrogynus; Sc, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe; Sj, Schizosaccharomyces japonicus; Ca, Candida albicans; An, Aspergillus nidulans; Mo, Magnaporthe oryzae; Nc, Neurospora crassa; Um, Ustilago maydis. See also Data S1, Data S2, Figure S2, and Figure S3.
We find that chytrids have both major classes of actin nucleators: formin family proteins that nucleate actin filaments de novo and the Arp2/3 complex, which typically nucleates a new actin filament on the side of an existing filament1,4. We find that each chytrid species has at least 4 formins with a diversity of domain organizations (described below). Each chytrid species also has at least one copy of each of the 7 Arp2/3 complex subunits and a variety of Arp2/3 activators (Figure S2). Some Arp2/3 activators are conserved in humans, chytrids, and Dikarya, such as WASP, and WISH/Dip1/SPIN90 (with the exception of Bs) (Figure S2). The SCAR/WAVE complex, in contrast, is present in humans and most chytrids (with the exception of Rg; Figure S2), but has been lost from the Dikarya10,16,17. The tight association between the SCAR/WAVE complex and cell migration suggests its retention correlates with cell migration10, implying that Rg may not have the capacity to crawl.
Actin is also subject to negative regulation, particularly by capping proteins that prevent further filament elongation and severing proteins that cut existing filaments. Many of these proteins are conserved in humans, chytrids, and Dikarya, such as capping proteins CapZ and AIP1, severing proteins Cofilin and Twinfilin, and the severing catalyst SRV2 (Figure 2). In contrast, the gelsolin/villin family of proteins is conserved in animals and chytrids, but not in most Dikarya (Figure 2). The conservation of these negative regulators indicates that Bd’s actin structures are likely dynamic.
We also identified a number of actin binding proteins conserved in humans, most chytrids, and Dikarya, including: Verprolin/WIP; Tropomyosin; Fimbrin/plastin; α-actinin; endo-/exocytosis proteins EPS15/Ede1, HIP1R/Sla2, and drebrin-like/ABP1 (Figure 2). We confirmed that while talin is not found in Dikarya, it is conserved in most chytrids18, with the interesting exception of Rg (Figure 2). Talin links adhesion receptors to the actin cytoskeleton in crawling cells19,20 and may be used during zoospore crawling, a hypothesis consistent with Rg’s lack of talin along with the SCAR/WAVE complex. Taken together, we find chytrid genomes encod a network of actin cytoskeletal regulators that is intermediate to the networks of animals and fungi. For an in-depth overview see Data S2.
Chytrids have typical fungal myosins as well as MyTH-FERM myosins
The myosin superfamily of actin-based motors has diverse cellular functions, including providing contractile forces during cell migration and cytokinesis, powering organelle transport, driving endocytosis, and building or maintaining actin-based structures such as filopodia. We searched for myosins in the same five species of chytrids and found myosins classes generally conserved among fungi: Myo1, Myo2, Myo5 and Myo17, a well as Myo22 (Figure S3, Data S2)21,22:
Myo1s are ancient, widely expressed myosins that link membranes to the actin cytoskeleton22,23. The budding and fission yeast Myo1s recruit activators of the Arp2/3 complex to actin patches to drive internalization of endocytic vesicles24. Each chytrid species has one or two Myo1s, which we predict may serve the same function and localize to the cortical actin patches observed in Bd and Sp zoospores and sporangia (Figure 1)2.
In contrast to the ubiquity of Myo1s, Myo2s are found mainly in Amoebozoans and Opisthokonts where they are an essential component of the cytokinetic contractile ring, generating forces necessary for the scission of two daughter cells during the final steps of mitosis25. Myo2s also play key roles in cell migration of animal cells and Amoebozoa, where they drive the retrograde flow of the actin network and generate cell polarity in migrating cells by contracting the actin network at the cell rear26. Chytrid fungi have a single Myo2 (Figure S3) that may play roles in zoospore crawling as well as cellularization (Figure 1).
Myosins also play key roles in intracellular transport, particularly Myo5s, which are present in Amoebozoa, Apusozoa, and Opisthokonts where they serve as actin-based transporters and localize cargo to the actin cortex27. Hyphal fungi use microtubules for long-distance transport and in these species Myo5s collaborate with kinesins. Many fungi have two Myo5s with distinct cellular functions. For example, one Myo5 of S. cerevisiae is required for organelle inheritance, polarized budding and mitotic spindle orientation while the second one is critical for polarized localization of cell fate determinants28. All five chytrid species contain at least one Myo5 that likely plays critical roles in intracellular transport, aiding in organelle segregation during division or targeting vesicles to sites of polarized growth.
Chytrids also have Myo17s. These unusual chimeric fungal myosins have a core motor domain arm fused to a chitin synthase 2 (Ch2) domain22. Transmembrane domains orient the protein so that upon exocytosis the chitin synthase enzyme is positioned toward the outer cell wall. Myo17s are essential for cell wall integrity and virulence in several pathogenic fungi29,30, and interestingly, a highly similar myosin is also found in molluscs31. Like other fungi, chytrids have multiple Myo17 family members that are likely used for targeted cell wall synthesis during different growth stages.
Although all chytrid species analyzed have at least one copy of Myo1, 2, 5, and 17, only some chytrid species appear to have retained Myo22. These myosins have one or two MyTH-FERM domains in the C-terminal tail region22 and are largely associated with the assembly and function of cellular protrusions composed of parallel bundles of actin, such as the filopodia of Dictyostelium and animal cells32–34. Although a subset of chytrid fungi including Rg and Sp have a single Myo22, neither Am, Bd nor Bs have a Myo22. We have not been able to detect these myosins in any other fungal species outside of the chytrids, suggesting that this myosin was lost early in fungal evolution.
Chytrid formins resemble those of fungi, animals, and plants, including DAAM and other diaphanous-related formins lost among the Dikarya
Formins play important roles in animal and fungal cell biology—formins nucleate the actin networks used for cytokinesis, cell movement, filopodia, and vesicle trafficking4. Although formins are defined by an FH2 domain that nucleates actin polymerization35–37, the biological function of each formin is heavily influenced by additional and highly variable protein domains that regulate its function and localization. Humans, for example, have 15 formins that are divided into seven main groups, four of which have a similar domain organization (diaphanous-related formins), while the remaining three have unique domain organizations 4. Although yeast formins share a similar domain organization to meta metazoan formins, yeast have far fewer formin genes—2 in S. cerevisiae and 3 in S. pombe4,38.
To investigate the possible roles formins might play in chytrid biology, we identified conserved domains in each chytrid formin using the Pfam database, and manually inspected protein sequences for the presence of FH1 domains and diaphanous autoregulatory domains. Here we describe the two most common domain organizations: Diaphanous-like formins and PTEN-like formins (for additional information about these and other chytrid formins, see Data S2).
The diaphanous-related formins have an N-terminal GTPase-binding domain (GBD) that binds to Rho GTPases to release the inhibitory interaction between the N-terminal inhibitory domain (FH3/DID) and the C-terminal diaphanous auto-regulatory domain (DAD)4,39. Each chytrid species contains at least one formin with a domain architecture that resembles that of diaphanous-related formins; containing a GTPase-binding domain, an FH3/DID domain, a coiled coil region, and an FH1 domain all N-terminal to the FH2 domain (Figure 3). As for the C-terminal diaphanous autoregulatory domain, only three proteins had the consensus sequence (MDXLLXXL)39, while the remaining proteins had similar sequences which may be functional if compensatory substitutions occurred in the FH3/DID (Figure 3, Data S3).
Figure 3. Chytrid formins share similar domain architectures to those of animals, Dikarya, and plants.
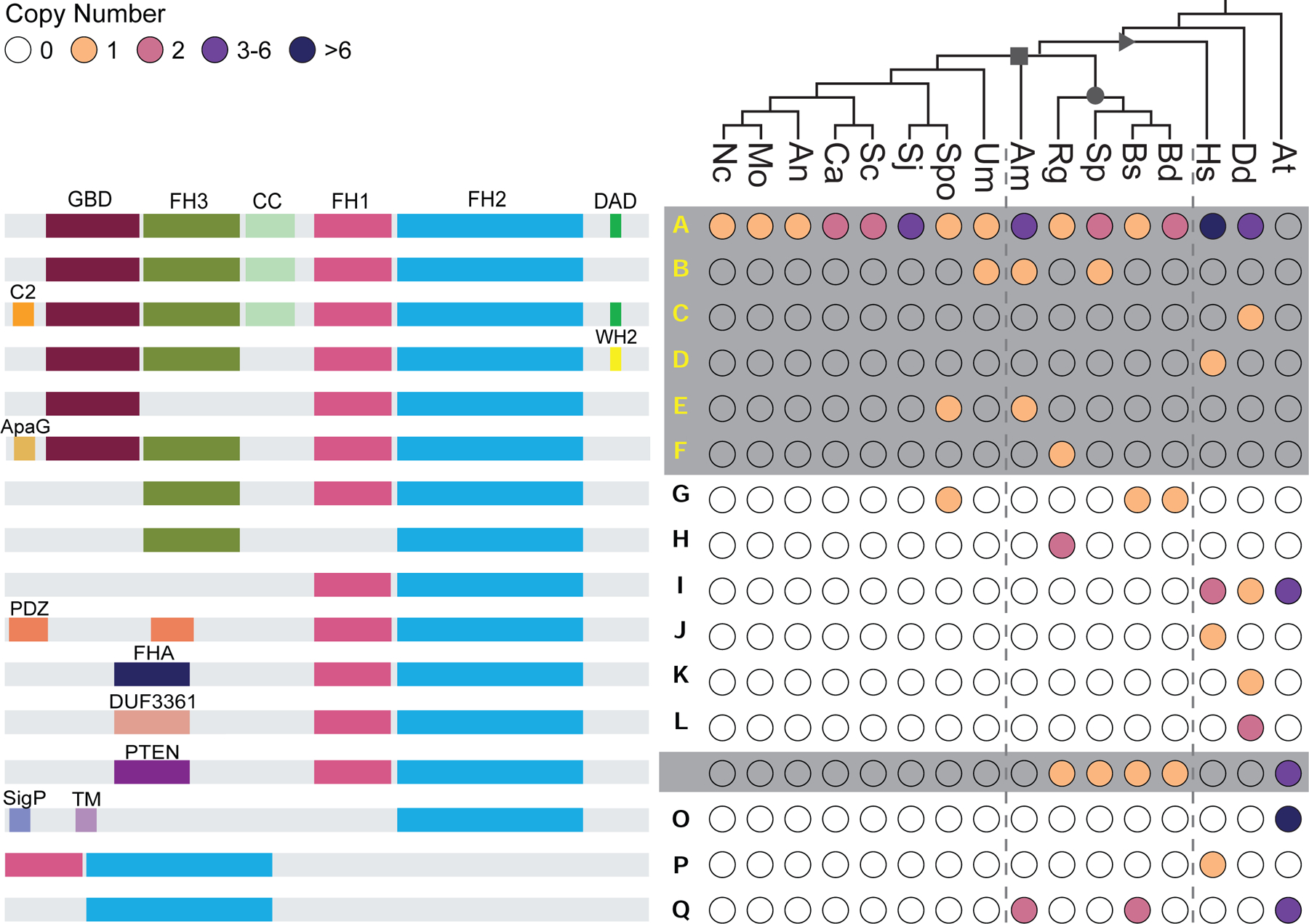
The distribution of given formin domain architectures (left, not to scale), across taxa (right). Each domain architecture is assigned a letter (middle) which is mapped onto Figure 4; yellow letters (A-F) indicate diaphanous-like architectures, white letters (M,N) indicate PTEN-domain-containing architectures (both plant and non-plant formins), black letters indicate architectures which did not fall into either of these classes. Color-filled circles (right) indicate the presence of at least one formin with the given domain architecture in that species, with color indicating the number of formins according to the key. Unfilled circles indicate that no formin with the indicated domain architecture was found in the given species. Symbols on the tree represent: opisthokonts (triangle); fungi (square); chytridiomycota (circle). Symbols in the circles represent: *, for at least one formin sequence in the indicated species, the DAD domain does not perfectly fit the consensus motif, but could potentially function as an autoregulatory domain. -, for at least one formin sequence, little to no sequence is present after the FH2 domain. #, although no region of this protein met the formal definition of an FH1 domain, a proline rich region (containing 4 polyproline stretches: 8 prolines/14 amino acids; 4/5; 4/6; and 7/11; total of 22 prolines over 197 amino acids) is found N-terminal to the FH2 domain in this protein (Genbank: ORY46833.1). ^, Dictyostelium formin ForC has no polyproline stretches and therefore no FH1 domain. At, Arabidopsis thaliana; Dd, Dictyostelium discoideum; Hs, Homo sapiens; Bd, Batrachochytrium dendrobatidis; Bs, Batrachochytrium salamandrivorans; Sp, Spizellomyces punctatus; Rg, Rhizoclosmatium globosum; Am, Allomyces macrogynus; Sc, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe; Sj, Schizosaccharomyces japonicus; Ca, Candida albicans; An, Aspergillus nidulans; Mo, Magnaporthe oryzae; Nc, Neurospora crassa; Um, Ustilago maydis. See also Data S3.
Each chytrid, except Am, also has a formin containing a PTEN/PTEN-like domain N-terminal to the FH2 domain (Figure 3). PTEN-formins are best known for their role in plants where they mediate membrane-localization by binding phospholipids40,41. Arabidopsis, for example, has 10 Class II formins, 4 of which have N-terminal PTEN-like domains that localize these formins to membranes4,40. The PTEN-like domain of class II formins has also been shown to mediate formin-microtubule interactions42.
Sharing domain organizations does not necessarily indicate relatedness, as the formin family is thought to have undergone gene duplication, divergence, and domain shuffling throughout its history38,41,43. We therefore wanted to determine how chytrid formins fit into this complex family history. We aligned 266 FH2 domain-containing sequences from 32 species across eukaryotic phyla to the Pfam FH2 domain (PF02181) full hidden Markov model, isolated the FH2-domains based on the predicted positions of the FH2 domain for S. cerevisiae Bni1p, and created a Maximum Likelihood phylogeny (Figure 4; Data S4, Data S5, Data S6).
Figure 4. Chytrids have animal-like formins related to DAAM a well as other diaphanous-related formins.
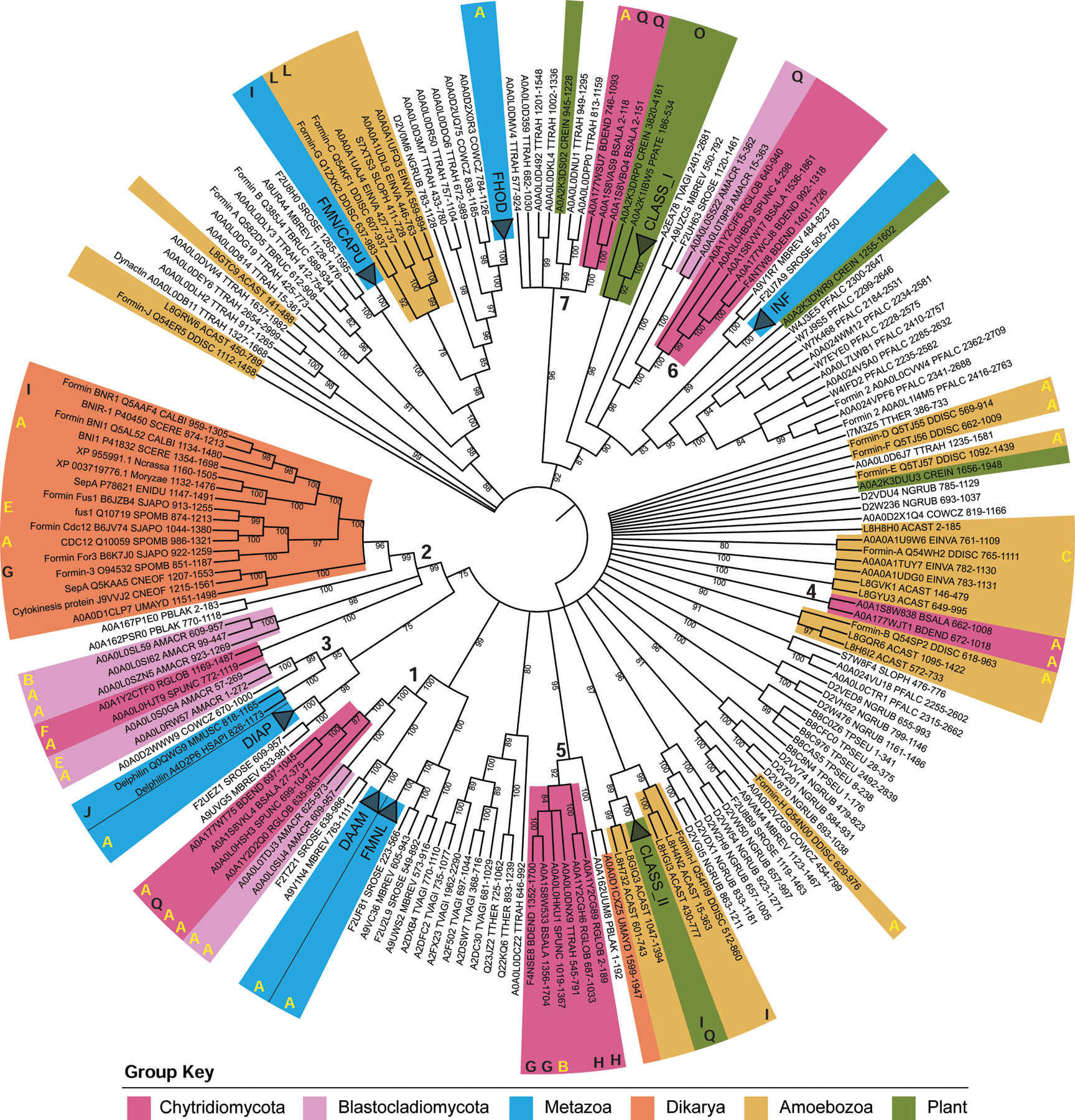
A maximum likelihood consensus tree was inferred using the FH2 domains of 266 formin proteins, rooted at the midpoint, with bootstrap values as shown, and nodes with <75% bootstrap support collapsed to polytomies. Metazoan clades and Arabidopsis thaliana clades were collapsed and named according to their formin group, except for Delphilin. Taxa of interest are colored according to the key, bold numbers indicate chytrid-containing clades. The bold letters around the outside of the tree correspond to the domain architectures in Figure 3; yellow letters indicate diaphanous-like architectures, white letters indicate PTEN-domain-containing architectures, black letters indicate architectures which did not fall into either of these classes. Protein names, Uniprot accession numbers, Full species name, Uniprot 5-letter species codes, position of the FH2 domain, and additional details can be found in Data S6. See also Data S4, Data S5, and Data S6.
Our phylogeny reveals that, surprisingly, most chytrid formins do not form a monophyletic group with other fungal formins, but instead are scattered throughout the tree in at least seven distinct clades (Figure 4), including: (1) DAAM-related formins with homologs in animals and their unicellular relatives; (2) bni1-type formins with homologs in other fungi; (3) Delphilin-related formins that have homologs in Allomyces but not in Chytridiomycetes; (4) a clade of formins found only in the parasitic chytrids Bd and Bs; (5) Fungi-2, a second clade of fungal formins known to be present in some fungi41; (6) non-plant PTEN/PTEN-like formins; and (7) a second Bd and Bs clade that also includes a homolog from the flagellated green algae Chlamydomonas. Proteins within each clade tend to share overall domain architectures. Interestingly, the chytrid PTEN/PTEN-like formins (Figure 4: architecture class M) are phylogenetically distinct from the plant Class II formins (Figure 4: architecture classes N, I, and Q), suggesting that their similarity in domain architecture may be due to convergent evolution41.
Bni1-type formins are associated with actin cables
Our phylogeny supports previous findings that yeast formins are evolutionarily distinct from animal formins (Figure 4, group 2)38,41,43. In budding yeast, Bni1p nucleates actin cables originating in the bud and Bnr1p nucleates actin cables primarily at the bud neck and into the mother cell44,45, while in fission yeast, the bni1-type formin For3 is responsible for nucleating cytoplasmic actin cables46. We find Bni-type formins in all other dikaryotic fungi included in our analyses (Figure 5A), as well as references to their assembly of actin cables47–50. We also find that some chytrids have at least one bni1-type formin, including Sp, whose sporangia we recently found can build actin cables within the cell body2. Other chytrids, including Bd and Bs, are missing these formins, an interesting finding given the lack of actin cables in the cell bodies of Bd zoospores and sporangia (Figure 1).
Figure 5. Bni1-type formins are associated with actin cables in the cell body.
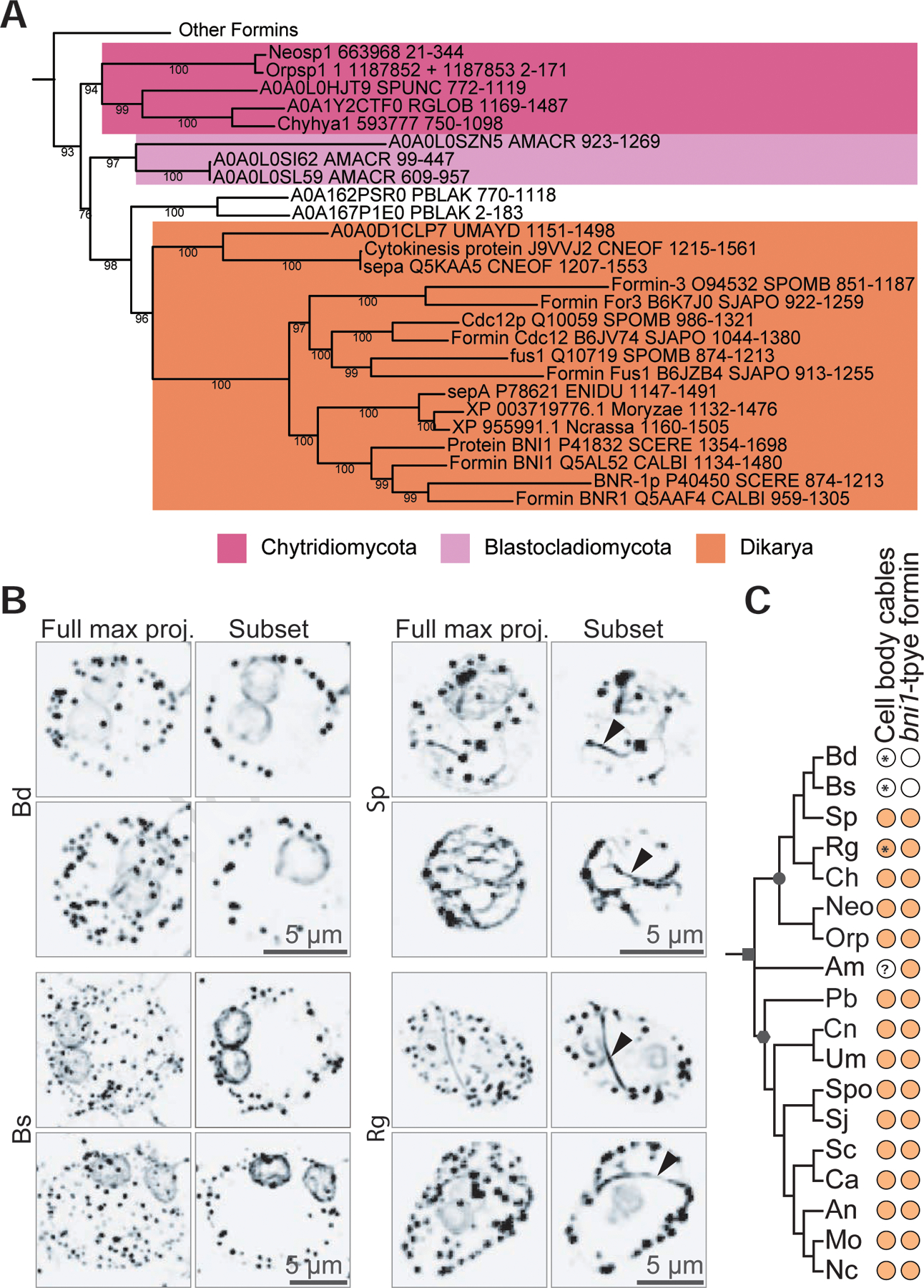
(A) The evolutionary history of 291 FH2 domain sequences from formin homologs was inferred by the maximum likelihood method for 349 amino acid positions in ≥80% of the sequences. This tree is the same as the tree in Figure 4, but includes the FH2 domains from the formins of three additional chytrid species (or their relatives) that have been observed to assemble actin cables in the cell body [Neocallimastix patriciarum51, Orpinomyces joyonii51, and Chytriomyces hyalinus52]. N. particiarum and O. joyonii did not have available genomes, so we used the genomes of species in the same genus [Neocallimastix californiae G165; Orpinomyces sp. strain C1A v1.066, assuming that the presence of cables is consistent across a genus. Consensus tree shown, pruned to highlight clade 2 (the main fungal clade) from Figure 4. Nodes with <75% bootstrap support were collapsed to polytomies. All bootstrap values are shown. (B) Representative examples of Batrachochytrium dendrobatidis (Bd), Batrachochytrium salamandrivorans (Bs), Spizellomyces punctatus (Sp), and Rhizoclosmatium globosum (Rg) sporangia stained for polymerized actin (inverted, black). Images are shown as a full maximum intensity projection as well as a subset of the z-stacks to highlight the middle sections of the cell body. Rg and Sp sporangia have actin patches, perinuclear actin shells, and actin cables within the cell body (arrowhead). Bd and Bs sporangia have actin patches and perinuclear actin shells, but no actin cables in the cell body. Scale bars, 5 µm. (C) Distribution of actin cables and bni1-type formins across fungi. Color-filled dots indicate the presence of the given component in the given species. Symbols on the tree represent: fungi (square); Dikarya (hexagon); chytridiomycota (circle). ? indicates the presence of cables in this species is unknown in the literature. * indicates a finding from this paper. Neo/Neosp1, Neocallimastix spp.; Orp/Orpsp1, Orpinomyces spp.; Bd/BDEND, Batrachochytrium dendrobatidis; Bs/BSALA, Batrachochytrium salamandrivorans; Sp/SPUNC, Spizellomyces punctatus; Rg/RGLOB, Rhizoclosmatium globosum; Ch/Chyhya1, Chytriomyces hyalinus; Am/AMACR, Allomyces macrogynus; Pb/PBLAK, Phycomyces blakesleeanus; Um/UMAYD, Ustilago maydis; Cn/CNEOF, Cryptococcus neoformans; An/ENIDU, Aspergillus nidulans; Sc/SCERE, Saccharomyces cerevisiae; Spo/SPOMB, Schizosaccharomyces pombe; Sj/SJAPO, Schizosaccharomyces japonicus; Ca/CALBI, Candida albicans; Mo/Moryzae, Magnaporthe oryzae; Nc/Ncrassa, Neurospora crassa.
We therefore hypothesized that a fungal species must have a bni1-type formin to build actin cables within the cell body. To e plore this hypothesis, we identified chytrid species that have been stained for actin, all of wh ch have actin cables within the cell body of their sporangia: Neocallimastix patriciarum 51; Orpinomyces joyonii51; and Chytriomyces hyalinus52, and added the formin FH2 domains from these species to the phylogeny to determine if they had at least one bni1-type formin. (Due to the lack of publicly available genome sequences for N. particiarum and O. joyonii, we used the genomes of species in the same genus: Neocallimastix californiae G1 and Orpinomyces sp. strain C1A). We find that all three additional putative cable-containing chytrid species have a bni1-type formin (Figure 5A). We also tested whether Bs, a chytrid with no bni1-type formin, has actin cables in its cell body and found no evidence of cables (Figure 5B). Finally, we fixed and stained Rg sporangia, as an example chytrid that has Bni-type formins but has not been observed to have cables. Similar to Sp, we find obvious cables within the cell body of Rg (Figure 5B). These findings are consistent with our hypothesis that bni1-type formins are used to build actin cables within the cell body of chytrid fungi that appear similar to the actin cables found in yeast and other dikaryotic fungi (Figure 5C).
Actin structures in Bd are dynamic and use distinct nucleators
To test our hypothesis that actin structures in Bd are dynamic, we treated Bd for short time periods with Latrunculin B (LatB), a specific small molecule inhibitor of actin polymerization that does not disrupt existing, stable filaments or networks. LatB treatment of zoospores revealed that the four actin phenotypes present in this life stage are dynamic, as the percent of cells displaying pseudopods, spikes, cortical actin, or patches decreased to nearly 0% with 30 minutes of treatment with LatB, compared to the ethanol carrier control (Figure 6).
Figure 6. Actin networks in Bd zoospores are dynamic and use distinct nucleators.
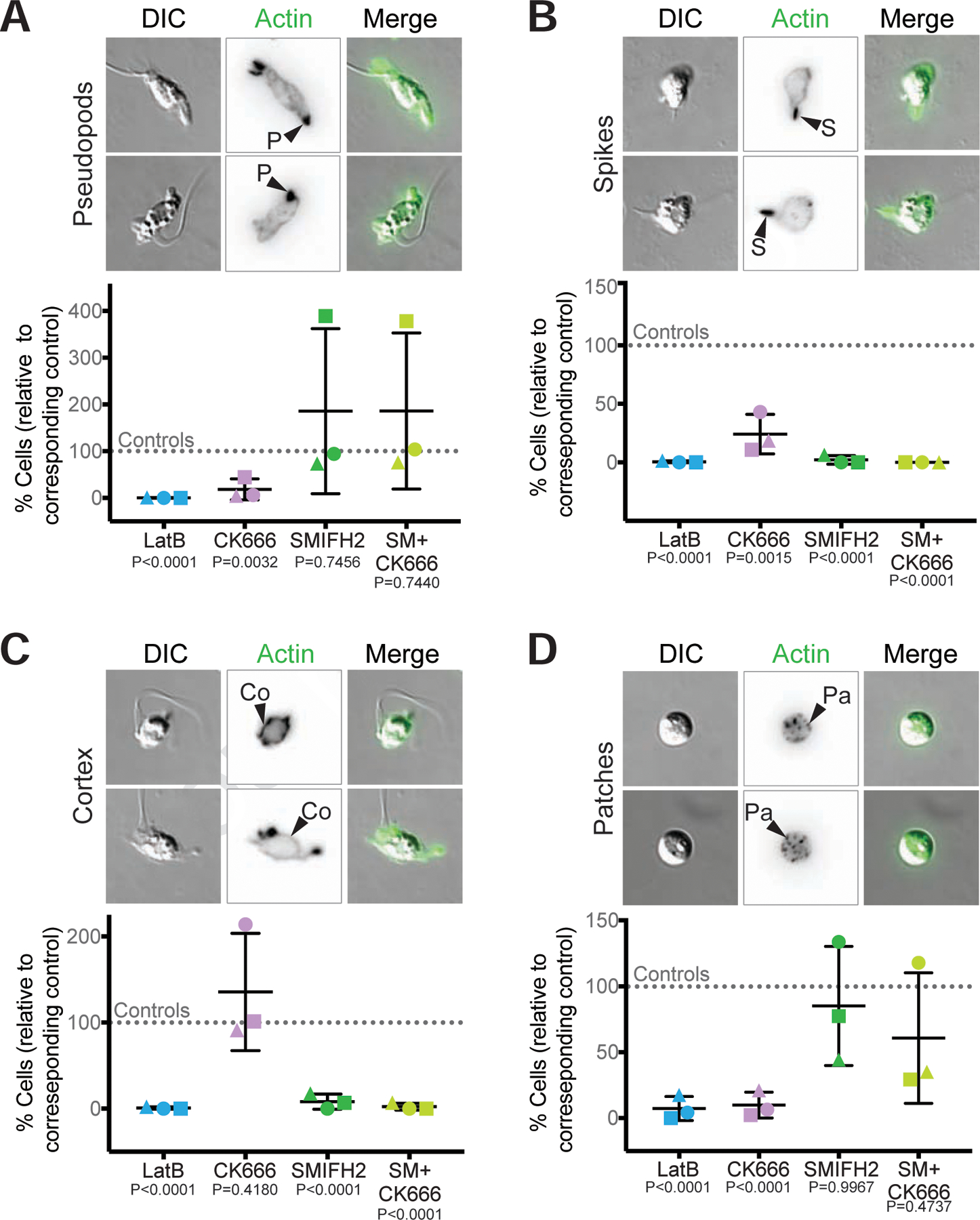
Synchronized populations of Bd zoospores were treated with LatrunculinB (LatB) to identify dynamic actin structures, or with Arp2/3 and/or formin inhibitors for 30 minutes. Cells were then fixed and stained for polymerized actin with fluorescent phalloidin, imaged, and quantified for presence of actin-filled pseudopods (P, part A), actin spikes (S, part B), cortical actin (Co, part C), and actin patches (Pa, part D). Each panel shows examples of cells (DIC: grey) and phalloidin-stained actin structures (alone inverted, black; overlay, green) with relative percent of cells with each structure quantified below. Percent of cells with the indicated actin phenotype for each drug treatment is normalized to its respective control: 1 µM LatrunculinB (LatB) normalized to the ethanol carrier control (EtOH); 100 µM CK666 normalized to its inactive analog CK689; 25 µM SMIFH2 normalized to a DMSO carrier control; and the combination treatment of 25 µM SMIFH2 + 100 µM CK666 (SM+CK666) was also normalized to the DMSO control. For raw data, see Data S7. Three independent experiments were performed, each represented by a different shape, the means and standard deviations shown in black. P-values for each treatment, relative to its respective control, are shown (unpaired Student’s T-tests for EtOH vs. LatB and CK689 vs. CK666; one-way ANOVA with Tukey’s multiple comparisons test for DMSO vs. SMIFH2 and the double treatment). Fluorescent images for (A), (B), and (D) are maximum intensity projections, fluorescent images for (C) are single z-slices to highlight the cortex. Brightness and contrast are not the same across images. Scale bars, 5 µm. See also Figure S4, Figure S5, and Data S7.
To determine which nucleators are required for building these dynamic actin structures, we treated zoospores and sporangia with CK666 that inhibits the Arp2/3 complex53,54, SMIFH2 that inhibits FH2 domains of formins55, or a combination of both. Chytrid Arp2/3 complexes (Figure S2C), as well as formin FH2 domains (Data S4) are similar to those from animal and fungal systems where these drugs have been previously validated53,55,56. SMIFH2 was recently shown to also affect some myosins57, so results from this drug could be due to either formin or myosin inhibition. For each treatment, we blindly measured the percentage of cells with each structure. Because of the variability in the prevalence of actin structures between biological replicates (Figure 1C, Data S7), as well as an effect of the drug carrier DMSO on actin phenotype frequencies (Figure S4), we normalized the data from each experimenta sample to its control (Figure 6, raw data in Data S7): 1 µM LatB normalized to the ethanol carrier control (EtOH), 100 µM CK666 normalized to its inactive analog CK689, 25 µM SMIFH2 normalized to a DMSO carrier control, and the combination treatment of 25 µM SMIFH2 + 100 µM CK666 (SM+CK666) was also normalized to the DMSO control. (Note: the results from normalizing the double treatment to the CK689 controls are equivalent to normalization with DMSO (Data S7). We then performed statistical tests on the normalized values and their respective controls, and inferred that each drug can penetrate cells sufficiently to induce phenotypes because each induced a large and statistically significant effect on at least one actin structure (Figure 6).
We first examined the role of Arp2/3 and formins in the assembly of zoospore actin structures. The percent of Bd zoospores with pseudopods decreased by an average of 80% with CK666 treatment (Figure 6A). SMIFH2 had a variable effect on pseudopods; in some trials, the drug had no impact, and in others it drastically increased the percent of cells with pseudopods (Figure 6A). The double treatment showed a similar pattern (Figure 6A). Although the protrusions in SMIFH2 treated zoospores fit our definition of a Bd pseudopod (actin-rich and at least 1 µm in width), the protrusions of the SMIFH2-treated cells appear rounder and less protrusive (Figure S5A). In contrast, all treatments decreased the percent of cells with actin spikes (Figure 6B). This effect, however, was less drastic with CK666 treatment alone, which decreased spikes by about 70% compared to about 98% for SMIFH2 and 100% for the double treatment (Figure 6B). While the effect of CK666 on cortical actin was variable (Figure 6C, Figure S1), the percent of cells with cortex consistently decreased by about 90% with SMIFH2 treatment and about 98% for the double treatment (Figure 6C). Treatment with CK666 reduced the percent of cells with ≥10 actin patches by an average of 90% (Figure 6D).
Next, we turned to sporangia and found that the perinuclear actin shells and actin patches have differing stability. Like those in zoospores, the actin patches in sporangia are highly dynamic and nearly disappear upon LatB treatment (8.3±6.3 versus 49.7±23.5 patches/cell in controls, Figures 7 and S6A). Actin patches in sporangia also appear to be Arp2/3-dependent as the average number of patches per cell decreased with CK666 treatment to 22.6±16.43 patches/cell, compared to 58±15.43 patches/cell in CK689 treated control cells (Figure 7B), an effect that was more pronounced in rhizoids (Figure 7C). Because CK666 treatment did not fully match treatment with LatB, we also measured the effects of the formin inhibitor SMIFH2 on actin patch number in sporangia (Figure 7). Treatment with SMIFH2 also reduced the average number of actin patches per cell (32.3±13.22), as did the double treatment (20±7.09 patches/cell) compared to the DMSO control (51.3±15.88 patches/cell). Interestingly, the location of the patches in the SMIFH2 treated cells remained relatively unchanged (Figure 7A), though for the double treatment, patches were severely reduced in the rhizoids (Figure 7C). Perinuclear shells appear more stable, as we observed no significant difference in the percent of nuclei encased by actin after treatment with actin inhibitors (Figure 7D). However, the intensity of the actin shells appeared asymmetric across the nucleus in some treatments. To quantitate this difference, we performed linescans and calculated the difference in the normalized intensity between each side. We found a slight increase in asymmetry with latrunculin and CK666 treatments, suggesting that the perinuclear actin shells may be only partially dynamic (Figures 7D, S6B). To assess the role of actin assembly in sporangial growth and maturation, we measured sporangial diameters after 24 and 48 hours of treatment (Figure S7A–D), as well as zoospore release (Figure S7E). LatB and CK666 treatment resulted in a clear reduction in both cell diameter and zoospore release, consistent with a requirement of actin polymerization in chytrid growth and development.
Figure 7. Actin patches in Bd sporangia are dynamic and use the Arp2/3 complex.
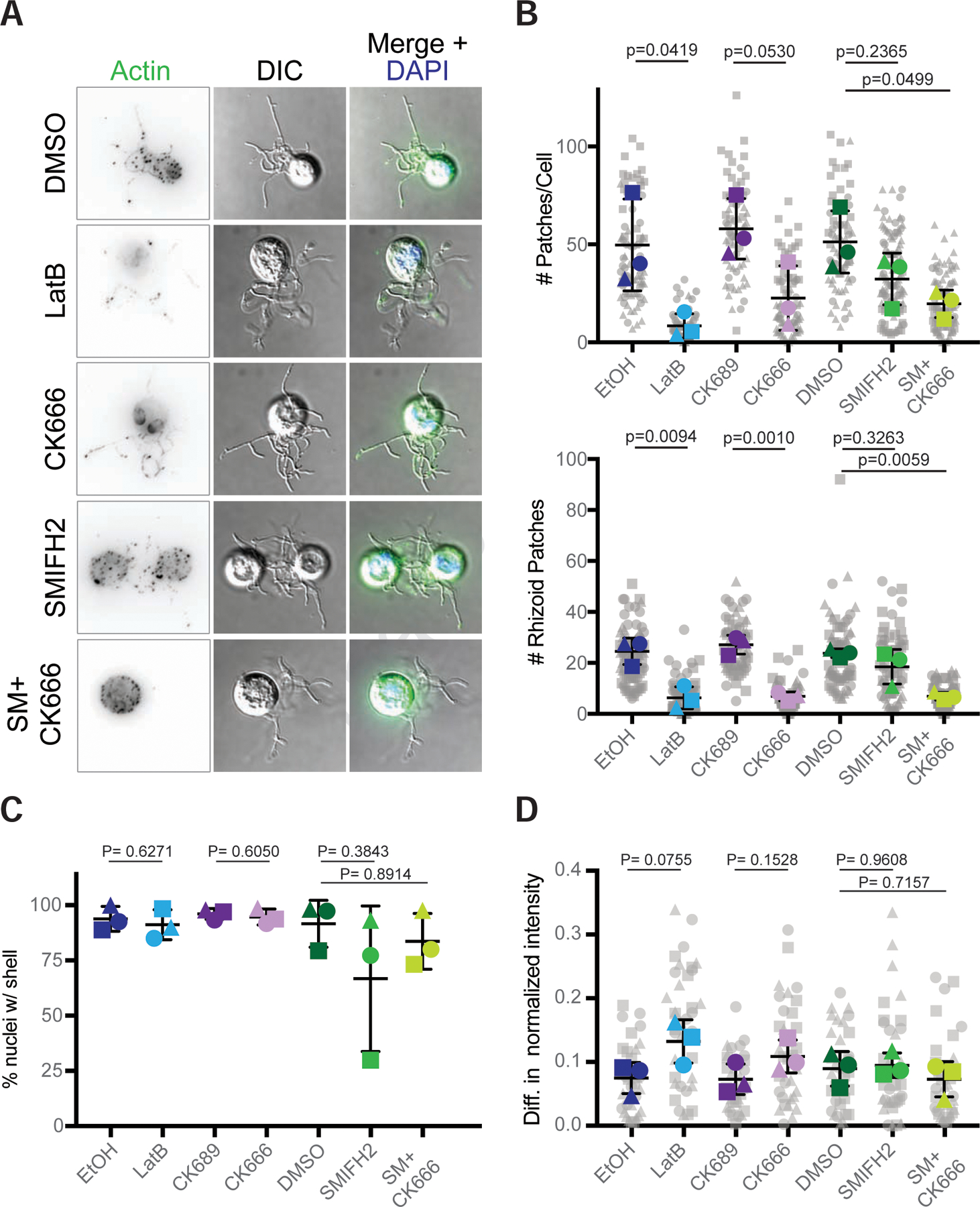
Populations of Bd sporangia seeded 1 day prior were treated with drugs using the same concentrations as in Figure 6, then fixed and stained for polymerized actin with phalloidin and for DNA with DAPI. (A) Examples of sporangia (DIC: grey) and phalloidin stained actin patches (inverted, black; green in overlay), with an overlay including the nucleus (blue) after treatment with each drug. Though DMSO has an effect on patches (see Figure S4), all controls looked phenotypically similar (see Figure S7), so only a DMSO treated cell is shown. (B) quantification of the number of actin patches per sporangia (top) and the number of patches in the rhizoids alone (bottom). Larger, colored shapes indicate the average number of patches per cell in each treatment from three independent experiments, each represented by a different shape. Each gray shape represents the number of patches in a single cell, or in a cell’s rhizoids in that experiment. Means and standard deviations of these averages are shown in black. (C) Percent of nuclei encased within an actin shell per treatment for three independent experiments. (D) Difference in the normalized intensity of actin shells on each side of the nucleus. The intensity along lines drawn through each nucleus were normalized to the center of each line. The average normalized intensity for each side was calculated, with the difference between the brightest half and the other half plotted here. Each gray shape represents the difference in normalized actin intensity for a single nucleus. Statistical tests were performed using the averages of the three experiments (i.e., the three colored shapes). P-values for each treatment, relative to its respective control, are shown (unpaired Student’s T-tests for EtOH vs. LatB and CK689 vs. CK666; one-way ANOVA with Tukey’s multiple comparisons test for DMSO vs. SMIFH2 and the double treatment). Brightness and contrast are not the same across images. Scale bar, 5 µm. See also Figure S6 and Figure S7.
These results show that most actin structures in Bd are dynamic and suggest that the Arp2/3 complex contributes to the formation of pseudopods and patches, while formins are used to build the cortex, and both appear to help build actin spikes.
DISCUSSION
Chytrids share a number of traits with other opisthokonts that are missing from Dikarya, including microtubule-based flagella58,59, cells that lack cell walls, and both animal-typical and fungal-typical cell cycle control machinery60. Here, we show that chytrid fungi also have an actin regulatory protein repertoire that appears intermediate to that of animals and Dikarya. For example, while animals, chytrids, and Dikarya all have a complete Arp2/3 complex, all members of the SCAR/WAVE complex, DAAM formins, gelsolin/villin family proteins, talin, and Myo22 (Figure 2, Figure S2) are conserved in animals and chytrid but are generally missing in the Dikarya. Other proteins thought to have been present in the last common eukaryotic ancestor are missing throughout the fungal lineage, such as the WASH complex and ENA/VASP family proteins (Figure 2, Figure S2) suggesting that they were lost very early during fungal evolution16,17. These findings highlight the potential for using chytrid fungi to explore actin cytoskeletal evolution.
Our analysis of formin evolution largely re apitulates the topologies reported in previous phylogenies38,41,43, but with greater taxonomic diversity and support and suggests that domain shuffling is common in the formins, particularly with PTEN/PTEN-like formins, which arose from at least two independent events41,61. This analysis also shows that, like other actin regulators, chytrids share formin families found in animals that are missing from Dikarya, including DAAM and Delphilin formins (Figure 4). Inclusion of multiple chytrid species in our analysis also revealed variability in formin content among chytrid species, including a remarkable correlation between the presence of bni1-type formins and fungal cells that build actin cables within their cell bodies (Figure 5). This suggests that bni1-type formins may be required specifically to build yeast-like actin cables. Bd, a species that lacks bni1-type formins, still builds spikes, pseudopods, cortical actin, and other actin structures, indicating that their assembly does not require bni1-type formins. The presence of ancestral formins alongside bni1-type formins would further suggest that cables evolved within organisms capable of building animal-like actin networks and were retained in Dikarya while the other formins were lost.
Bd zoospores assemble a variety of dynamic actin structures whose appearance and regulation appear similar to those of human cells. In addition to Arp2/3-dependent pseudopods (Figure 6A)10, Bd zoospores build thin, actin-filled protrusions we call spikes. Like the thin, actin-filled filopodia of animal cells that require both Arp2/3 and formin activity1, spikes in Bd are sensitive to both CK666 and SMIFH2 (Figure 6). Although shorter than animal (on average 5–20 µm5) and Dictyostelium filopodia (2–5 µm62), the visual similarity as well as the apparent involvement of both Arp2/3 complex and formin protein activity, raises the possibility that these actin spikes are related to filopodia.
While the zoospore actin cytoskeleton resembles that of human cells, the actin cytoskeleton of Bd sporangia more closely resembles that of yeast and other Dikarya, particularly in the assembly of small actin patches near the cell periphery (Figure 1D). Based on their similarity to actin patches of yeast and other Dikarya, we predict that these actin patches are involved in endocytosis, explaining their abundance in growing sporangia. We hypothesize that the patches seen in a minority of zoospores represent cells that have initiated their transition to the sporangial growth stage (Figure 1B) an idea consistent with their increased cell wall staining and circularity (Figure S5B–D). Like actin patches of yeast, the actin patches of Bd sporangia are sensitive to Arp2/3 inhibition (Figure 7B). Bd sporangia also build perinuclear actin shells (Figure 1D), similar to those found in other species of chytrid fungi. Earlier reports of perinuclear shells suggested they were fixation artifacts51, but recent live imaging of actin in Sp clearly shows that these structures are present in living sporangia and form just before mitosis2.
We know of no Dikarya cells with the animal-like actin phenotypes seen in chytrid zoospores. Chytrids use these structures for crawling, a behavior not seen in sessile fungi that spend their life cycle enclosed in cell walls. The simplicity of the actin cytoskeleton in Dikarya, therefore, may have corresponded with the loss of the zoospore stage type that actin uses these actin regulators to build animal-like structures.
In addition to important clues about the evolution of Dikarya studying chytrid actin networks provides us with valuable information about chytrid biology. Bd is a causative agent of Chytridiomycosis, a deadly skin infection of amphibians that is associated with population declines around the world13,58, and the actin structures we see could play important roles in the infection process. For example, zoospores could use pseudopods to crawl along the surface of the host to find a suitable local environment before encysting. Spikes, if they function like filopodia, could also be used for movement or for sensing local environmental conditions. Additionally, actin patches likely facilitate nutrient uptake to fuel sporangial growth and production of new zoospores, a finding consistent with the observed role of actin polymerization in sporangial growth and maturation (Figure S7). This model suggests that actin networks underlie the motility and rapid growth that are key to the pathology and pathogenicity of Bd.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lillian Fritz-Laylin (lfritzlaylin@umass.edu)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All data are available in the figures, tables, and data files associated with this manuscript. This study did not result in any unique code.
Experimental model and subject details
Cultures of Bd JEL423 were maintained in 1% Tryptone broth at 24°C in non-ventilated, tissue-culture treated flasks. For experiments investigating zoospores, populations seeded 3 days prior were synchronized to ensure the cells were in a similar developmental stage. Synchronization was achieved by washing out previously-released zoospores 3 times with 1% Tryptone and adding fresh Tryptone for the sporangia left adhered to the flask. The sporangia were incubated at 24°C for 2 hours before newly released zoospores were harvested from suspension via centrifugation at 2000xg for 5 minutes. For experiments investigating sporangia, cells from populations seeded in non-tissue culture treated flasks 24-hours 1 day prior (called 1-day cultures) were harvested from suspension via centrifugation at 2000xg for 5 minutes.
Cultures of Batrachochytrium salamandrivorans (Bsal) isolate AMFP 1 were grown in half-strength TGhL liquid media (0.8% Tryptone, 0.2% gelatin hydrolysate, 0.1% lactose (w/v)). For fixing and staining sporangia, cells were seeded in non-tissue culture-treated vented flasks, grown for 48-hours (2-day cultures), and harvested via centrifugation at 2000xg for 5 minutes.
Cultures of Spizellomyces punctatus (Koch type isolate NG-3) Barr (ATCC 48900) were grown and harvested as in2. Briefly, cells were grown in Koch’s K1 liquid medium67 (1L; 0.6 g peptone, 0.4 g yeast extract, 1.2 g glucose, 15 g agar if plates) at 30°C, transferred to K1 agar plates at room temperature, and harvested by flooding the plate with dilute salt (DS) solution68 48 hours later and collected by centrifugation.
Cultures of Rhizoclosmatium globosum JEL800 were grown on K1 Penicillin/streptomycin plates (10ml/L: Thermo-Fisher P4333–20ML) at 23°C. Zoospores were harvested by flooding the plate with 1 mL DS and filtered with a sterile syringe filter with Whatman paper #1. Zoospores were counted in a neubauer chamber and diluted with DS to 1x106 zoospores/mL. Then 200 uL of the diluted zoospores were added to a glass-bottomed 8-well imaging dish and incubated for 10 minutes at 23°C. The DS solution was carefully removed, the well was washed once with K1 Penicillin/streptomycin liquid media, then replaced with fresh K1 Penicillin/streptomycin liquid media. The 8-well dish was left to incubate for ~22 hours at 23°C inside a plastic petri dish sealed with parafilm. Cells were fixed and stained directly in the imaging dish, see below for details.
Methods Details
Identification of actin regulatory proteins
Protein sequences from the following chytrids were retrieved from the website of the National Center for Biotechnology Information (NCBI; ncbi.nlm.nih.gov): Batrachochytrium dendrobatidis (Bd) strains JAM81 (GCA_000203795.1, JGI-PGF project ID 4001669; http://genome.jgi-psf.org/Batde5) and JEL42369, Batrachochytrium salamandrivorans69 (Bs; GCA_002006685.1); Spizellomyces punctatus DAOM BR11770 (Sp; GCA_000182565.2); Rhizoclosmatium globosum JEL80071 (Rg; GCA_002104985.1); and Allomyces macrogynus ATCC 38327 (Am; GCA_000151295.1, Broad Institute). Homologs of proteins from the model organisms used in this study [Saccharomyces cerevisiae S288C (Sc), Schizosaccharomyces pombe strain 972 (Spom), Schizosaccharomyces japonicus strain yFS275 (Sj), Candida albicans strain SC5314 (Ca), Aspergillus nidulans strain FGSC A4 (An), Magnaporthe oryzae str in 70–15 (Mo), Neurospora crassa strain OR74A (Nc), and Ustilago maydis strain 512 (Um), Dictyostelium discoideum AX2 and AX4 (Dd), Homo sapiens, Mus musculus (occasionally used to confirm results), and Arabidopsis thaliana (At)] were identified using a combination of the Basic Local Alignment Search Tool72 (BLAST), literature review, and probing Swiss-Prot reviewed entries on UniProtKB. Individual UniProtKB IDs or NCBI/Genbank accession numbers for proteins from these model species are provided in Data S2, along with the NCBI accession numbers for the proteins from t indicated chytrid species. Additional sequences were retrieved from NCBI from species used for actin identification [Oryctolagus cun culus, Gi rdia intestinalis (GIAIN), Chlamydomonas reinhardtii (CHLRE)]. Multiple splice variants were not included in this analysis.
Actin:
BLASTp using standard parameters (E= 1.0x10−5, word size=3, BLOSUM 62 matrix, filtering low complexity regions) and Rabbit, Sc, Spom, and Dd actin sequences as queries was used to identify actin homologs in the five chytrid species of interest and the remaining Dikarya species. Reciprocal BLASTs and domain analysis were not used for identifying actin sequences because there are many paralogues and highly similar actin-related proteins which make determining a mutual best BLAST hit (MBBH) difficult. Instead, sequences sharing ≥50% sequence identity with any of thes queries were compiled and aligned with the query sequences as well as actin sequences from GIAIN and CHLRE; Arp1 from human, Sc, Spom, and Dd; and Arp2 and Arp3 from human, Sc, Spom, Dd, and the five chytrids (Data S1). Given that most actin sequences are highly conserved across species, the 50% threshold chosen for gathering potential actin sequences is very low for this protein and likely caught all potential actin sequences. With the alignment, a simple Maximum Likelihood phylogeny was built using the IQtree web server73 (default parameters). The phylogeny was rooted at the Arp3 clade, and branches with less than 70% bootstrap support were collapsed to polytomies. Chytrid and other Dikarya actin sequences were defined as those which formed a clade with known actin sequences.
Actin regulatory proteins:
BLASTp with standard parameters (E= 1.0x10−5, word size=3, BLOSUM 62 matrix, and filtering low complexity regions) and queries from Sc, Spom, Dd, mouse, or human was used to identify homologs for 34 actin regulatory proteins, complexes, or protein families in the five chytrid species of interest and the remaining Dikarya. BLAST hits were confirmed by obtaining a MBBH and hits were further confirmed by identifying the predicted domain structure of all potential chytrid or Dikarya homologs. MBBHs were found through NCBI (same parameters) or yeastgenome.org74 (TBLASTN, default parameters, open reading frames dataset) using the potential chytrid or Dikarya homologs as search queries. The predicted domains of all potential chytrid or Dikarya homolog sequences were obtained using the hmmscan tool (hmmer.org) against the Pfam database75 (v32) using the website of the European Bioinformatics Institute (EMBL-EBI; https://www.ebi.ac.uk/) or on the command line using hmmscan from the hmmer suite v3.2.1 (hmmer.org) and the PfamA Hidden Markov Model75 (v32). In some cases, MBBHs were not obtained due to the complexity of the protein family, but domain architectures and multiple sequence alignments of the chytrid homologs confirm their family membership.
Myosin:
Full length myosin protein sequences for Allomyces macrogynus (ATCC 38327), Batrachochytrium dendrobatidis (JAM81), Spizellomyces punctatus (DAOM BR117) Saccharomyces cerevisiae (RM11–1a), Schizosaccharomyces pombe (972h), Schizosaccharomyces japonicus (yFS275), Candida albicans (SC5314), Aspergillus nidulans (FGSC A4), Neurospora crassa (OR74A), and Ustilago maydis (512), Arabidopsis thaliana, Dictyostelium discoideum (AX4) and Homo sapiens were obtained from Cymobase76 (www.cymobase.org). Myosin sequences for Batrachochytrium salamandrivorans, Rhizoclosmatium globosum and Magnaporthe oryzae (70–15) were identified by extensive BLAST searches using either full-length, myosin motor domain or MyTHFERM domain from Sp Myo22 as a query. The classification of the myosins was validated by a general BLAST search of the full protein and manual inspection of the tail domains.
Identification of FH2 domain-containing proteins
Identifying potential formin sequences required a different a proach from the rest of the actin regulatory proteins. Reciprocal BLAST hits were not used to identify formin family proteins because of the complexity of the family both within and across species. Formins were gathered by similarity to known formins and then confirmed as a formin based on the presence of an FH2 domain. We used the amino acid sequences corresponding to the predicted FH2 domains from Sc formins Bni1p and Bnr1p as independent search queries using Position Specific Iteration-BLAST77 (PSI-BLAST) through NCBI (default parameters, filter low complexity regions). For each chytrid species and for each search query, five iterations of PSI-BLAST were run, after which convergence occurred and subsequent iterations yielded no new sequences above the E-value threshold (0.005). All sequences above the threshold after these iterations were checked for an FH2 domain using the domain prediction method from above; those without an FH2 domain were removed from the dataset. Surrounding gene annotations of proteins with only FH2 domains were checked to identify other formin-typical domains to create a full length formin sequence. Splice variants were not included in this analysis. For the Dikarya species that are not Sc or Spom, BLASTp with standard parameters (E= 1.0x10−5, word size=3, BLOSUM 62 matrix, and filtering low complexity regions) and full length formin queries from Sc, Spom, Human, Dd, and At were used. The top 5 hits for each query in each species were gathered and checked for the presence of an FH2 domain. Sequences without an FH2 domain were removed from the dataset.
Identification of formin domain organizations
All known formin sequences from human, Sc, Spom, Dd, and At were checked for predicted domains using the same domain prediction method as above and confirming these results with the literature. All chytrid and remaining dikaryotic FH2-domain containing sequences were checked for predicted domains as well. Coiled coil regions, signal peptides (SigPs) in plant formins, and transmembrane (TMs) in plant formins were determined using this method, as EBI runs hmmscan, a coiled coil prediction algorithm78, and the Phobius program79 for SigPs and TMs simultaneously. FH1 domains contain many polyproline stretches and are hard to determine through computational methods, thus FH1 domains in chytrid and the remaining dikaryotic sequences were determined by hand. Polyproline stretches were defined as being at least 6 prolines long out of 7 consecutive residues, the minimum number of prolines needed to bind profilin80–83. An FH1 domain was defined as the stretch of amino acids from the first proline of the first polyproline stretch to the last proline of the last polyproline stretch directly N-terminal to the FH2 domain. The diaphanous autoregulatory domain of diaphanous like formins is short and also needed to be identified by hand. This domain is C-terminal to the FH2 domain and has the consensus sequence MDXLLXXL39. The C-termini of all chytrid and remaining dikaryotic formins were aligned using TCoffee (default parameters) and checked for the consensus sequence (Data S3). Sequences with obvious insertions or deletions in the diaphanous autoregulatory domain region were removed and the remaining sequences were realigned and reexamined.
Phylogenetic analysis of formin proteins
Main analysis:
FH2-domain-containing sequences were obtained from the species distribution sunburst of the FH2 domain page (PF02181) on the Pfam website75 (v32; pfam.xfam.org). The following species in the given taxa were used in this analysis: Chytridiomycota: Bd, Bs, Sp, Rg; Blastocladiomycota: Am; Mucoromycota: Phycomyces blakesleeanus; Dikarya: Sc, Spom, Aspergillus nidulans, Cryptococcus neoformans, Ustilago maydis, Schizosaccharomyces japonicus, Candida albicans, Magnaporthe oryzae, and Neurospora crassa; Microsporidia: Spraguea lophii; Metazoa: Human, mouse, Drosophila melanogaster; Choanoflagellates: Monosiga brevicollis, Salpingoeca rosetta; Filasterea: Capsaspora owczarzaki; Apusozoa: Thecamonas trahens; Amoebozoa: Dd, Acanthamoeba castellanii, Enantoboeba invadens; Discoba: Naegleria gruberi, Trypanosoma brucei; SAR: Thalassiosira pseudonana, Plasmodium falciparum, Tetrahymena thermophila; Plants: At, CHLRE, Physcomitrella patens; Metamonads: Trichomonas vaginalis. The CD-hit program84,85 was used to remove sequences which were ≥98% identical, reducing redundancy in the data set. The remaining sequences were aligned to the full HMM for the FH2 domain from Pfam using hmmalign (no additional program options) from the hmmer suite v3.2.1. The FH2 domains were isolated by trimming the alignment according to the FH2 domain of the Sc Bni1p sequence (starting PHKKLKQ; ending ADFINEY), which had been previously hand clipped and provided an accurate judgement for the placement of other FH2 domains. Columns which had <80% occupancy were removed from the alignment (Data S4, Data S6). A Maximum Likelihood phylogeny (Data S5, Data S6) was generated from this curated alignment using the IQTree webserver73 (default parameters).
bni1-type formin analysis:
We searched the literature and identified the following chytrid species with observed actin cables: Neocallimastix patriciarum51; Orpinomyces joyonii51; and Chytriomyces hyalinus52. N. particiarum and O. joyonii did not have genomes in the JGI database, so we used the genomes of species in the same genus, assuming that the presence of cables is consistent across a genus. We used the FH2 domains from Bd JAM81 formins as queries for TBLASTN searches (default settings, perform gapped alignments) against the filtered model transcripts database for the following species’ genomes on JGI MycoCosm: Neocallimastix californiae G165; Orpinomyces sp. strain C1A v1.066; Chytriomyces hyalinus JEL632 v1.0 (JGI, https://mycocosm.jgi.doe.gov/Chyhya1/). All hits were confirmed for the presence of an FH2 domain using the same method described above. FH2-domain-containing sequences from these three species were added to the file containing the formin sequences used in the main analysis and the same alignment, clipping, column editing, and tree building processes were performed as before. The large tree was pruned using the iTOL website86 (v5.5.1) to show only the main fungal clade that included Bni1p and Bnr1p from Sc.
Chemical inhibition of actin and actin nucleators
Synchronized Bd zoospores were adhered to the bottom of 96-well plates using 0.5 mg/mL Concanavalin A (Sigma, C2010). Adhered cells were then treated with Bonner’s Salts87, 1uM LatrunculinB (Millipore, 4280201MG) or an equal volume of ethanol, 100uM CK666 (Calbiochem/Sigma, 182515) or 100uM of the inactive analog CK689 (Calbiochem/Sigma, 182517), 25uM SMIFH2 (Tocris Bioscience, 440110) or equal volumes of DMSO, and 100uM CK666 + 25uM SMIFH2 for 30 minutes. Cells were fixed using fixation buffer (4% PFA and 50 mM Sodium Cacodylate, pH=7.2) on ice for 20 minutes, permeabilized and stained for DNA using a mixture of 0.1% Triton X and DAPI (0.5 µg/ml; Life Technologies, D1306) in PEM buffer (100mM PIPES, 1mM EGTA, 0.1mM MgSO4) at room temperature for 10 minutes, and then stained for polymerized actin using Alexa Fluor488 Phalloidin (0.2 U/mL; Life Technologies, A12379) in PEM buffer at room temperature for at least one hour. For experiments investigating the correlation between cell shape and actin patches, cells were not stained with DAPI, and instead cell walls were stained with 0.1% Calcofluor White for 10 minutes and washed out before permeabilization of the cells with Triton.
Sporangia from 1-day Bd and Sp cultures and from 2-day Bsal cultures were adhered to the bottom of 96-well plates using ~0.1% Polyethyleneimine (Sigma, P3143) and then treated, fixed, and stained using the same procedure used for Bd zoospores with phalloidin and DAPI. For long term drug treatments on Bd sporangia, synchronized zoospores were seeded into three tissue-culture treated 24-well plates at a density of ~2x105 cells/mL in 1% Tryptone broth. These cells grew for 24 hours at 24°C, after which the media was removed and 1 day cells with no drugs added to them were stained for the cell wall with 0.1% Calcofluor White. The remaining two plates did not have the media removed, but had drugs added to them in the same concentrations as done for the short-term experiments. These cells then grew at 24°C for either another 24 hours (for 2-day cells that have been treated with drugs for 24 hours) or 48 hours (for 3-day cells that have been treated with drugs for 48 hours). Plates with 2-day cells and 3-day cells that have been treated with drugs for the indicated times were stained for the cell wall. Zoospore release per treatment was counted 72-hours after initial seeding of cells using a Burker Turk hemocytometer.
Rg sporangia ~22 hours into development, were fixed and stained in the imaging dish they were grown in. These cells were fixed with fixation buffer (4% PFA and 50 mM Sodium Cacodylate, pH=7.2) on ice for 20 minutes, then treated with 50 µg/mL chitinase (Sigma, C6137–5UN) in 20 mM potassium phosphate buffer (pH= 6.0) for 1 hour at room temperature before permeabilizing and staining as for the other chytrid species.
Bd zoospores were categorized based on the actin structures present in each cell: pseudopods, actin spikes, cortical actin, actin patches, or a combination of any of these (Data S7).
Microscopy
For the short-term drug experiments, cells were imaged on an inverted microscope (Ti-2 Eclipse; Nikon) with a 100X 1.45 NA oil objective and using NIS Elements software. Images were taken using both differential interference contrast (DIC) microscopy and widefield fluorescence microscopy with 460 nm to visualize phalloidin and 360 nm light to visualize DAPI. For the long-term drug experiments, cells were imaged on an inverted microscope (Ti-2 Eclipse; Nikon) with a 40X 0.06 NA air objective in DIC and fluorescence microscopy with 360 nm light to visualize the cell wall stained with Calcofluor White. For imaging sporangia for the cable analysis, chytrid sporangia were imaged on an inverted microscope (Ti-2 Eclipse; Nikon) with a 100X 1.45 NA oil objective using NIS Elements software. Images were taken using both differential interference contrast (DIC) microscopy and spinning-disc confocal fluorescence microscopy with 460 nm to visualize phalloidin and 360 nm light to visualize DAPI. All images were taken in Z-stacks to encompass the whole cell. All imaging was done at room temperature in PEM buffer.
Quantification and statistical analysis
Widefield image processing and analysis was performed in Fiji88, and blind scoring was performed using the CellCounter FIJI plugin (Kurt De Vos, https://imagej.nih.gov/ij/plugins/cell-counter.html). Confocal images were deconvolved using Autoquant X3 software with default settings (10 iterations, medium noise, expert recommended settings), and a full maximum intensity projection image of the actin staining was created as well as a subset projection image to highlight the presence or absence of actin cables in Bd, Bsal, Sp, and Rg cell bodies.
Bd zoospores were categorized based on the actin structures present in each cell: pseudopods, actin spikes, cortical actin, actin patches, or a combination of any of these (Data S7). Pseudopods were defined as bright areas of actin staining 1–2 µm wide, while spikes were defined as being less than 1 µm wide and at least 1 µm long. To analyze cortex intensity, we drew a box extending from outside of the cell into the cell and measured the average phalloidin intensity along that box in a representative area of the cell. Cortical actin was defined as bright staining for actin along the outer edge of at least 50% of the cell. While it misses some information, this method was chosen over including the whole cortica region to exclude obvious actin patches, which often localize to the cell edge, and which would skew the data. Actin patches were defined as at least 10 bright spots of actin <1 µm in diameter in the cell.
The number of patches in each Bd sporangium was counted and patch position (cell body vs. rhizoid) was noted. Perinuclear shells, defined as rings of actin around each nucleus in a sporangium, were quantified by counting the number of nuclei and the number of actin shells in every cell for each treatment. The intensity of these shells was also quantified by line scans 5 pixels in width. At least 10 nuclei per treatment were randomly chosen. Lines were drawn across a representative area across roughly the center of the nucleus and avoiding actin patches as much as possible. The percent of nuclei with actin shells was calculated for each drug treatment for three independent exp riments. For the shell line scans, the lines were normalized both by percent along the line [(distance along the line/total line length)*100)] and by intensity to the average middle intensity for each line. To make visualizing the data easier, the data was ordered such that the most intense value for each line was on the side. The intensities for data points from 0–49.99% (left side of the line) and from 50–100% (right side of the line) were averaged for each line. The difference between the normalized intensities on the left and the normalized intensities on the right side of each line were calculated by subtraction and averaged for each of the three biological replicates.
For zoospores, the percent of cells with each actin structure for each treatment was normalized to its respective control: LatrunculinB (LatB) normalized to the ethanol carrier control (EtOH), CK666 normalized to its inactive analog CK689, SMIFH2 normalized to a DMSO carrier control, and the combination treatment SMIFH2 + CK666 (SM+CK666) was also normalized to the DMSO control. We chose to normalize the SM+CK666 data to DMSO rather than to CK689 because the data more closely resemble the SMIFH2 data, making it easier to present. Normalization to CK689 instead of DMSO does not change the results of the experiments (Data S7). Statistical tests were performed on these normalized values and their respective control, for the three independent experiments. To determine relationships between encystment, cell shape, and actin patches in Bd zoospores, the outline of control cells focused at the coverslip was traced using the kidney bean tool in FIJI with a line width of 1 pixel in the phalloidin channel. Outlines included any protrusions from the cell. Then, with the outline still drawn, the average intensity of cell wall staining within the outline and the circularity value of the outline were determined. The average calcofluor white intensity and circularity values for cells with and without actin patches for three independent replicates were calculated. The individual values for cells with and without actin patches were also plotted for intensity vs. circularity for three independent experiments. For the short-term drug experiments in Bd sporangia, the average number of patches per cell, and in each cell’s rhizoids in each treatment was calculated for three independent experiments.
For the long-term drug treatments, Calcafluor White stained sporangia were thresholded in NIS elements using the following parameters: “per channel”; smooth 5x; clean 5x; fill holes on; separate 3x; with a size range of 5–380um except for samples treated with LatB, CK666, and SM+CK666 which used a size range of 3–380 um. Size ranges varied between treatments to account for large differences in cell size due to the effects of the drugs. Once a binary layer was created for each image, the objects were counted using the “count objects” function in NIS Elements. Binary objects which extended past the field of view, contained more than one sporangium, included more than half of the rhizoid area, or did not fill at least half of the sporangium were removed from analysis. Because the sporangia are roughly spherical, the EqDiameter metric was used to estimate the sporangial diameters for each treatment by calculating the diameter of a sphere with equal volume. The concentratio of released zoospores after 72 hours of growth (48 hours of treatment) was coun ed usin a Burker Turk hemocytometer. These data were normalized as for the short-term zoosp re drug treatments.
Statistical tests were performed using the averages of the three experiments. For all statistical tests, we used unpaired Student’s T-tests for EtOH vs. LatB and CK689 vs. CK666 and a one-way ANOVA with Tukey’s multiple comparisons test for DMSO vs. SMIFH2 and the double treatment.
Supplementary Material
Chytrid and remaining Diakrya sequences sharing ≥50% sequence identity with actin from rabbit, budding yeast, fission yeast, or Dictyostelium were compiled and aligned with these actin sequences as well as actin sequences from Giardia intstialis (GIAIN) and Chlamydomonas reinhardtii (CHLRE); Arp1 from human, budding and fission yeast, and Dictyostelium; and Arp2 and Arp3 from human, budding and fission yeast, Dictyostelium and the five chytrids. TCoffee was used to align the sequences (default parameters). The alignment was not edited, and was used for Maximum Likelihood phylogeny estimation (see Figure S2A). The beginnings of NCBI/Genbank accession numbers for chytrid sequences are as follows: XP_0066, Batrachochytrium dendrobatidis (Bd) JAM81; OAJ, Bd JEL423; OON, Batrachochytrium salamandrivorans; XP_0166, Spizellomyces punctatus; ORY, Rhizoclosmatium globosum; KNE, Allomyces macrogynus. Non-chytrid sequences are labeled with the Uniprot accession number (if applicable) and the protein and organism name except for Arp2 in Saccharomyces cerevisiae (“ARP2”) and Schizosaccharomyces pombe (“SPAC11H11.06”). The new dikarya sequences are the last 7 accession numbers. For chytrid and new Dikarya actin accession numbers see Figure S2 and Data S2.
The raw data from scoring actin phenotypes in Batrachochytrium dendrobatidis zoospores. Pa, patches; S, spike; P, pseudopod; C, cortex. NP, nondescript protrusion. Note: NP was used in our preliminary study design using lower resolution imaging methods. The final data sets were higher resolution and allowed for the distinction between pseudopods and spikes in all cases, resulting in zero “NP” cells. “Uninterpretable” cells are cells for which we were not able to accurately assess phenotypes because they were either clumped in groups, partially out of the frame, or out of focus; these cells were excluded from the analysis. “Haze” indicates cells that had no distinct actin structures, but were positive for phalloidin staining. The SMIFH2+CK666 treatment was normalized to both appropriate controls: DMSO as the control for SMIFH2 or CK689 as the control for CK666. Normalizing to one or the other did not affect the results and both are provided in this file.
The C-terminus of each chytrid formin was hand clipped directly after the end of the FH2 domain, and aligned using TCoffee (default parameters). Sequences with obvious insertions in the diaphanous autoregulatory domain (DAD) region (for reference: starting at XP_016609770.1 M58 (M1240 in the full protein), or position 350 in the alignment) were removed and remaining sequences were realigned as before and evaluated for the presence of the consensus sequences for a DAD (MDXLLXXL)S50.
FH2-domain-containing sequences were obtained from the species distribution sunburst of the FH2 domain page (PF02181) on the Pfam websiteS51 (v32; pfam.xfam.org). Sequences which were ≥98% identical were removed to reduce redundancy and the remaining sequences were aligned to the full HMM for the FH2 domain from Pfam. The FH2 domains were isolated by trimming the alignment according to the FH2 domain of the Saccharomyces cerevisiae Bni1p sequenc (starting PHKKLKQ; ending ADFINEY), which had been previously hand clipped and provided an accurate judgement for the placement of other FH2 domains. Columns which had <80% occupancy were removed from the alignment. This file was used to create the main formin phylogeny in this paper (see Figure 4 and Data S5).
The raw data in newick format of the formin phylogeny in Figure 4. Built by maximum likelihood method using the alignment in Data S4 with default parameters on the IQtree webserverS52.
Protein identifiers given by Pfam upon sequence download were renamed for species with cryptic identifiers before continuing analysis. This file contains the information used for protein identifiers in Figure 4, Figure 5, Data S4 and Data S5, as well as some additional information. The “PFAM_name” column indicates the original identifier of the sequence when downloaded. The “UNIPROT_ACC” column indicates the un prot accession number for the sequence. The “organism_name” column gives the name of the species the sequence is from, “Orgnaism_code” gives the species’ Uniprot 5-letter species code, “Organism_strain” indicates the specific strain of the organism the sequence is from. The “Gene_name” column indicates the name of the protein used in the phylogeny. The “ORF_range” indicates the amino acid position of the FH2 domain in the full protein sequence.
On the first sheet are additional details about the actin cytoskeletal components found in chytrids and other species. The second sheet has the genbank/NCBI accession numbers for the homologs of actin and actin regulatory proteins in the given chytrid species. Chytrid homologs were identified using BLAST with queries from several species of Dikarya, humans, Dictyostelium discoideum, and Arabidopsis thaliana; uniprot accession numbers given. On the third sheet are the genbank/NCBI and Uniprot accession numbers for the homologs of myosin motor proteins in the given chytrid and Dikarya species, humans, Dictyostelium discoideum, and Arabidopsis thaliana.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Penicillin/streptomycin | Thermo-Fisher | Cat#P4333–20ML |
| Concanavalin A | Sigma | Cat#C2010 |
| LatrunculinB | Millipore | Cat#4280201MG |
| CK666 | Calbiochem/Sigma | Cat#182515 |
| CK689 | Calbiochem/Sigma | Cat#182517 |
| SMIFH2 | Tocris Bioscience | Cat#440110 |
| DAPI | Life Technologies | Cat#D1306 |
| Alexa Fluor488 Phalloidin | Life Technologies | Cat#A12379 |
| Polyethyleneimine | Sigma | Cat#P3143 |
| Chitinase | Sigma | Cat#C6137–5UN |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Batrachochytrium dendrobatidis strain JEL423 | Joyce Longcore | JEL423 |
| Batrachochytrium salamandrivorans isolate AMFP1 | Frank Pasmans | AMFP1 |
| Spizellomyces punctatus Koch type isolate NG-3 | ATCC | 48900 |
| Rhizoclosmatium globosum strain JEL800 | Tim James (https://czeum.herb.lsa.umich.edu/) | JEL800 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Basic Local Alignment Search Tool (BLAST) | 72 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Position Specific Iteration-BLAST (PSI-BLAST) | 77 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| UniprotKB | https://www.uniprot.org/ | N/A |
| HMMER suite v3.2.1 | Hmmer.org | Hmmer.org |
| Pfam database v32 | 75 | pfam.xfam.org |
| European Bioinformatics Institute (EMBL-EBI) | https://www.ebi.ac.uk/ | https://www.ebi.ac.uk/ |
| Coiled Coiled predictor | 78 | N/A |
| Phobius program | 79 | N/A |
| CD-hit program | 84,85 | N/A |
| IQTree webserver | 73 | http://iqtree.cibiv.univie.ac.at/ |
| iTOL v5.5.1 | 86 | https://itol.embl.de/ |
| FIJI | 88 | https://imagej.net/Fiji/Downloads |
| CellCounter FIJI plugin | Kurt De Vos | https://imagej.nih.gov/ij/plugins/cell-counter.html |
| Autoquant X3 vX3.1.3 | Media Cybernetics | https://www.mediacy.com/79-products/autoquant-x3 |
| NIS Elements w/ Advanced Research Package | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| Other | ||
| Batrchochytirum dendrobatidis JAM81 reference assembly v1.0 | US DOE Joint Genome Institute | NCBI: GCA_000203795.1 |
| Batrachochytirum dendrobatidis JEL423 assembly | Broad Institute | NCBI: GCA_000149865.1 |
| Batrachochytrium salamandrivorans representative assembly (assembly Batr_sala_BS_V1) | Broad Institute | NCBI: GCA_002006685.1 |
| Spizellomyces punctatus DAOM BR117 representative assembly (S_punctatus_V1) | Broad Institute | NCBI: GCA_000182565.2 |
| Rhizoclosmatium globosum JEL800 representative assembly (Rhihy1) | DOE Joint Genome Institute | NCBI: GCA_002104985.1 |
| Allomyces macrogynus ATCC 38327 representative assembly (A_macrogynus_V3) | Broad Institute | NCBI: GCA_000151295.1 |
| Saccharomyces cerevisiae S288C NCBI RefSeq build R64 assembly | Saccharomyces Genome Database | NCBI: GCF_000146045.2 |
| Schizosaccharomyces pombe strain 972 NCBI RefSeq assembly | S. pombe European Sequencing Consortium (EUPOM) | NCBI: GCF_000002945.2 |
| Schizosaccharomyces japonicus strain yFS275 NCBI RefSeq assembly | Broad Institute | NCBI: GCF_000149845.2 |
| Candida albicans strain SC5314 NCBI RefSeq assembly (assembly ASM18296v3) | Stanford University | NCBI: GCA_000182965.3 |
| Aspergillus nidulans strain FGSC A4 NCBI RefSeq assembly (assembly ASM14920v2) | Broad Institute | NCBI: GCA_000149205.2 |
| Magnaporthe oryzae strain 70–15 NCBI RefSeq assembly (Pyricularia oryzae 70–15 assembly MG8)) | International Rice Blast Genome Consortium | NCBI: GCA_000002495.2 |
| Neurospora crassa strain OR74A NCBI RefSeq assembly (assembly NC12) | Broad Institute | NCBI: GCA_000182925.2 |
| Ustilago maydis strain 512 NCBI RefSeq assembly (assembly Umaydis521_2.0) | Broad Institute | NCBI: GCA_000328475.2 |
| Dictyostelium discoideum AX4 NCBI Reference assembly (assembly dicty2.7) | The Dictyostelium discoideum Sequencing Consortium | NCBI: GCA_000004695.1 |
| Human reference genome on NCBI (assembly GRCh38.p13) | Genome Reference Consortium | NCBI: GCA_000001405.28 |
| Mus musculus reference genome on NCBI (assembly GRCm39) | Genome Reference Consortium | NCBI: GCF_000001635.27 |
|
Arabidopsis thaliana reference genome on NCBI
(assembly TAIR10.1) |
The Arabidopsis Information Resource | NCBI: GCA_000001735.2 |
| Oryctolagus cuniculus reference genome on NCBI (assembly OryCun2.0) | The Genome Sequencing Platform, The Assembly Computation and Development Core Team | NCBI: GCA_000003625.1 |
| Giardia intestinalis representative genome on NCBI (assembly UU_WB_2.1) | Marine Biological Laboratory | NCBI: GCA_000002435.2 |
| Chlamydomonas reinhardtii representative genome on NCBI (assembly v3.0) | DOE Joint Genome Institute | NCBI: GCA_000002595.2 |
| Neocallimastix californiae G1 genome | 51 | https://mycocosm.jgi.doe.gov/Neosp1/Neosp1.home.html |
| Orpinomyces sp. strain C1A v1.0 genome | 51 | https://mycocosm.jgi.doe.gov/Orpsp1_1/Orpsp1_1.home.html |
| Chytriomyces hyalinus JEL632 v1.0 genome | 52 | https://mycocosm.jgi.doe.gov/Chyhya1/ |
Highlights.
Chytrid fungi diverged before the radiation of Dikarya (multicellular fungi & yeast)
Chytrids have actin genes and structures typical of both Dikarya and animal cells
The regulation of chytrid actin structures resembles that of animal/dikaryotic cells
Presence of a BNI1/BNR1 type formin correlates with the presence of actin cables
ACKNOWLEDGEMENTS
We thank Tom Pollard, Harry Higgs, and Jessica Henty-Ridilla for feedback on Figure 2, as well as Madelaine Bartlett, Edgar Medina, Katrina Velle, and Samuel Lord for comments on the manuscript. We also thank Katrina Velle for assisting in deconvolution of confocal images, Edgar Medina for assisting with growth of Spizellomyces punctatus and Rhizoclosmatium globosum, and Terrell Redwood and Tatihana Beckford for assisting with data analysis. This material is based upon work supported by the Pew Charitable Trust (to L.K.F.-L.) and the National Institutes of Health (R01GM12291 to M.A.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Blanchoin L, Boujemaa-Paterski R, Sykes C, and Plastino J (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev 94, 235–263. [DOI] [PubMed] [Google Scholar]
- 2.Medina EM, Robinson KA, Bellingham-Johnstun K, Ianiri G, Laplante C, Fritz-Laylin LK, and Buchler NE (2020). Genetic transformation of Spizellomyces punctatus, a resource for studying chytrid biology and evolutionary cell biology. elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velle KB, and Fritz-Laylin LK (2019). Diversity and evolution of actin-dependent phenotypes. Curr. Opin. Genet. Dev 58–59, 40–48. [DOI] [PubMed] [Google Scholar]
- 4.Breitsprecher D, and Goode BL (2013). Formins at a glance. J. Cell Sci 126, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallop JL (2020). Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell Dev. Biol 102, 81–89. [DOI] [PubMed] [Google Scholar]
- 6.Clark AG, Dierkes K, and Paluch EK (2013). Monitoring actin cortex thickness in live cells. Biophys. J 105, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haase K, and Pelling AE (2013). Resiliency of the plasma membrane and actin cortex to large-scale deformation. Cytoskeleton (Hoboken) 70, 494–514. [DOI] [PubMed] [Google Scholar]
- 8.Moseley JB, and Goode BL (2006). The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev 70, 605–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudin O, Bendezú FO, Groux R, Laroche T, Seitz A, and Martin SG (2015). A formin-nucleated actin aster concentrates ce ll wall hydrolases for cell fusion in fission yeast. J. Cell Biol 208, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz-Laylin LK, Lord SJ, and Mullins RD (2017). WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. J. Cell Biol 216, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burianek LE, and Soderling SH (2013). Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol 24, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossart H-P, Van den Wyngaert S, Kagami M, Wurzbacher C, Cunliffe M, and Rojas-Jimenez K (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol 17, 339–354. [DOI] [PubMed] [Google Scholar]
- 13.O’Hanlon SJ, Rieux A, Farrer RA, Rosa GM, Waldman B, Bataille A, Kosch TA, Murray KA, Brankovics B, Fumagalli M, et al. (2018). Recent Asian origin of chytrid fungi causing global amphibian declines. Science 360, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. (2006). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822. [DOI] [PubMed] [Google Scholar]
- 15.Berger L, Hyatt AD, Speare R, and Longcore JE (2005). Life cycle stages of the amphibian chytrid Batrachochytrium dendrobatidis. Dis. Aquat. Org. 68, 51–63. [DOI] [PubMed] [Google Scholar]
- 16.Veltman DM, and Insall RH (2010). WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell 21, 2880–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollmar M, Lbik D, and Enge S (2012). Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res. Notes 5, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebé-Pedrós A, Roger AJ, Lang FB, King N, and Ruiz-Trillo I (2010). Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci USA 107, 10142–10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niewöhner J, Weber I, Maniak M, Müller-Taubenberger A, and Gerisch G (1997). Talin-null cells of Dictyostelium are strongly defective in adhesion to particle and substrate surfaces and slightly impaired in cytokinesis. J. Cell Biol 138, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priddle H, Hemmings L, Monkley S, Woods A, Patel B, Sutton D, Dunn GA, Zicha D, and Critchley DR (1998). Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol 142, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odronitz F, and Kollmar M (2007). Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kollmar M, and Mühlhausen S (2017). Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol 17, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntosh BB, and Ostap EM (2016). Myosin-I molecular motors at a glance. J. Cell Sci 129, 2689–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giblin J, Fernández-Golbano IM, Idrissi F-Z, and Geli MI (2011). Function and regulation of Saccharomyces cerevisiae myosins-I in endocytic budding. Biochem. Soc. Trans 39, 1185–1190. [DOI] [PubMed] [Google Scholar]
- 25.West-Foyle H, and Robinson DN (2012). Cytokinesis mechanics and mechanosensing. Cytoskeleton (Hoboken) 69, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguilar-Cuenca R, Juanes-García A, and Vicente-Manzanares M (2014). Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell. Mol. Life Sci 71, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titus MA (2018). Myosin-Driven Intracellular Transport. Cold Spring Harb. Perspect. Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui Y (2003). Polarized distribution of intracellular components by class V myosins in Saccharomyces cerevisiae. Int. Rev. Cytol 229, 1–42. [DOI] [PubMed] [Google Scholar]
- 29.Takeshita N, Yamashita S, Ohta A, and Horiuchi H (2006). Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol 59, 1380–1394. [DOI] [PubMed] [Google Scholar]
- 30.Gandía M, Harries E, and Marcos JF (2014). The myosin motor domain-containing chitin synthase PdChsVII is required for development, cell wall integrity and virulence in the citrus postharvest pathogen Penicillium digitatum. Fungal Genet. Biol 67, 58–70. [DOI] [PubMed] [Google Scholar]
- 31.Weiss IM, Schönitzer V, Eichner N, and Sumper M (2006). The chitin synthase involved in marine bivalve mollusk shell formation contains a myosin domain. FEBS Lett 580, 1846–1852. [DOI] [PubMed] [Google Scholar]
- 32.Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, and Titus MA (2001). A role for myosin VII in dynamic cell adhesion. Curr. Biol 11, 318–329. [DOI] [PubMed] [Google Scholar]
- 33.Petersen KJ, Goodson HV, Arthur AL, Luxton GWG, Houdusse A, and Titus MA (2016). MyTH4-FERM myosins have an ancient and conserved role in filopod formation. Proc Natl Acad Sci USA 113, E8059–E8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weck ML, Grega-Larson NE, and Tyska MJ (2017). MyTH4-FERM myosins in the assembly and maintenance of actin-based protrusions. Curr. Opin. Cell Biol 44, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, and Boone C (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- 36.Sagot I, Rodal AA, Moseley J, Goode BL, and Pellman D (2002). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol 4, 626–631. [DOI] [PubMed] [Google Scholar]
- 37.Kovar DR, Kuhn JR, Tichy AL, and Pollard TD (2003). The fis ion yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol 161, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalkia D, Nikolaidis N, Makalowski W, Klein J, and Nei M (2008). Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol 25, 2717–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgs HN (2005). Formin proteins: a domain-based approach. Trends Biochem. Sci 30, 342–353. [DOI] [PubMed] [Google Scholar]
- 40.Grunt M., Zárský V., and Cvrcková F. (2008). Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol 8, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruyne D (2017). Probing the origins of metazoan formin diversity: Evidence for evolutionary relationships between metazoan and non-metazoan formin subtypes. PLoS ONE 12, e0186081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kollárová E, Baquero Forero A, Stillerová L, Přerostová S, and Cvrčková F (2020). Arabidopsis class II formins atfh13 and atfh14 can form heterodimers but exhibit distinct patterns of cellular localization. Int. J. Mol. Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgs HN, and Peterson KJ (2005). Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruyne D, Gao L, Bi E, and Bretscher A (2004). Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buttery SM, Yoshida S, and Pellman D (2007). Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell 18, 1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feierbach B, and Chang F (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol 11, 1656–1665. [DOI] [PubMed] [Google Scholar]
- 47.Delgado-Álvarez DL, Bartnicki-García S, Seiler S, and Mouriño-Pérez RR (2014). Septum development in Neurospora crassa: the septal actomyosin tangle. PLoS ONE 9, e96744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda T, Takaine M, Numata O, and Nakano K (2016). Anillin-related protein Mid1 regulates timely formation of the contractile ring in the fission yeast Schizosaccharomyces japonicus. Genes Cells 21, 594–607. [DOI] [PubMed] [Google Scholar]
- 49.Li Y-B, Xu R, Liu C, Shen N, Han L-B, and Tang D (2020). Magnaporthe oryzae fimbrin organizes actin networks in the hyphal tip during polar growth and pathogenesis. PLoS Pathog 16, e1008437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, Loh ZY, Xue J, Zhou F, Sun J, Qiao Z, Jin S, Deng Y, Li H, Wang Y, et al. (2020). Orchestrated actin nucleation by the Candida albicans polarisome complex enables filamentous growth. J. Biol. Chem [DOI] [PMC free article] [PubMed]
- 51.Li J, and Heath IB (1994). The Behavior of F-Actin during the Zoosporic Phases of the Chytridiomycete Gut Fungi Neocallimastix and Orpinomyces. Exp. Mycol 18, 57–69. [Google Scholar]
- 52.Dee JM, Landry BR, and Berbee ML (2019). Actin guides filamentous rhizoid growth and morphogenesis in the zoosporic fungus Chytriomyces hyalinus. Mycologia 111, 904–918. [DOI] [PubMed] [Google Scholar]
- 53.Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, and Pollard TD (2009). Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hetrick B, Han MS, Helgeson LA, and Nolen BJ (2013). Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activa ing conformational change. Chem. Biol. 20, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, and Kovar DR (2009). Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol 16, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, and Kovar DR (2014). Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol 24, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura Y, Shi S, Zhang F, Liu R, Takagi Y, Bershadsky AD, viasnoff, virgile, and Sellers JR (2020). The formin inhibitor, SMIFH2, inhibits members of the myosin superfamily. BioRxiv. [DOI] [PMC free article] [PubMed]
- 58.Longcore JE, Pessier AP, and Nichols DK (1999). Batrachochytrium Dendrobatidis gen. et sp. nov., a Chytrid Pathogenic to Amphibians. Mycologia 91, 219. [Google Scholar]
- 59.Hodges ME, Scheumann N, Wickstead B, Langdale JA, and Gull K (2010). Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci 123, 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medina EM, Turner JJ, Gordân R, Skotheim JM, and Buchler NE (2016). Punctuated evolution and transitional hybrid network in an ancestral cell cycle of fungi. elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gisbergen PAC, Wu S-Z, Chang M, Pattavina KA, Bartlett ME, and Bezanilla M (2018). An ancient Sec10-formin fusion provides insights into actin-mediated regulation of exocytosis. J. Cell Biol 217, 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, and Gerisch G (2007). Organization of actin networks in intact filopodia. Curr. Biol 17, 79–84. [DOI] [PubMed] [Google Scholar]
- 63.Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, and Trask BJ (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet 3, e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghoshdastider U, Popp D, Burtnick LD, and Robinson RC (2013). The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton (Hoboken) 70, 775–795. [DOI] [PubMed] [Google Scholar]
- 65.Haitjema CH, Gilmore SP, Henske JK, Solomon KV, de Groot R, Kuo A, Mondo SJ, Salamov AA, LaButti K, Zhao Z, et al. (2017). A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol 2, 17087. [DOI] [PubMed] [Google Scholar]
- 66.Youssef NH, Couger MB, Struchtemeyer CG, Liggenstoffer AS, Prade RA, Najar FZ, Atiyeh HK, Wilkins MR, and Elshahed MS (2013). The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol 79, 4620–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koch WJ (1968). Studies of the motile cells of chytrids. v. flagellar retraction in posteriorly uniflagellate fungi. Am. J. Bot. 55, 841–859. [Google Scholar]
- 68.Machlis L (1953). Growth and nutrition of water molds in the subgenus euallomyces. i. growth factor requirements. Am. J. Bot 40, 189–195. [Google Scholar]
- 69.Farrer RA, Martel A, Verbrugghe E, Abouelleil A, Ducatelle R, Longcore JE, James TY, Pasmans F, Fisher MC, and Cuomo CA (2017). Genomic innovations linked to infection strategies across emerging pathogenic chytrid fungi. Nat. Commun 8, 14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russ C, Lang BF, Chen Z, Gujja S, Shea T, Zeng Q, Young S, Cuomo CA, and Nusbaum C (2016). Genome Sequence of Spizellomyces punctatus. Genome Announc 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, et al. (2017). Widespread adenine N6-methylation of active genes in fungi. Nat. Genet 49, 964–968. [DOI] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 73.Trifinopoulos J, Nguyen L-T, von Haeseler A, and Minh BQ (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44, W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Odronitz F, and Kollmar M (2006). Pfarao: a web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase). BMC Genomics 7, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lupas A, Van Dyke M, and Stock J (1991). Predicting coiled coils from protein sequences. Science [DOI] [PubMed]
- 79.Käll L, Krogh A, and Sonnhammer ELL (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol 338, 1027–1036. [DOI] [PubMed] [Google Scholar]
- 80.Perelroizen I, Marchand JB, Blanchoin L, Didry D, and Carlier MF (1994). Interaction of profilin with G-actin and poly(L-proline). Biochemistry 33, 8472–8478. [DOI] [PubMed] [Google Scholar]
- 81.Petrella EC, Machesky LM, Kaiser DA, and Pollard TD (1996). Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35, 16535–16543. [DOI] [PubMed] [Google Scholar]
- 82.Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, and Almo SC (1999). Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol 6, 666–671. [DOI] [PubMed] [Google Scholar]
- 83.Paul AS, and Pollard TD (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Y, Niu B, Gao Y, Fu L, and Li W (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Letunic I, and Bork P (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonner JT (1947). Evidence for the formation of cell aggregates by chemotaxis in the development of the slime mold Dictyostelium discoideum. J. Exp. Zool 106, 1–26. [DOI] [PubMed] [Google Scholar]
- 88.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chytrid and remaining Diakrya sequences sharing ≥50% sequence identity with actin from rabbit, budding yeast, fission yeast, or Dictyostelium were compiled and aligned with these actin sequences as well as actin sequences from Giardia intstialis (GIAIN) and Chlamydomonas reinhardtii (CHLRE); Arp1 from human, budding and fission yeast, and Dictyostelium; and Arp2 and Arp3 from human, budding and fission yeast, Dictyostelium and the five chytrids. TCoffee was used to align the sequences (default parameters). The alignment was not edited, and was used for Maximum Likelihood phylogeny estimation (see Figure S2A). The beginnings of NCBI/Genbank accession numbers for chytrid sequences are as follows: XP_0066, Batrachochytrium dendrobatidis (Bd) JAM81; OAJ, Bd JEL423; OON, Batrachochytrium salamandrivorans; XP_0166, Spizellomyces punctatus; ORY, Rhizoclosmatium globosum; KNE, Allomyces macrogynus. Non-chytrid sequences are labeled with the Uniprot accession number (if applicable) and the protein and organism name except for Arp2 in Saccharomyces cerevisiae (“ARP2”) and Schizosaccharomyces pombe (“SPAC11H11.06”). The new dikarya sequences are the last 7 accession numbers. For chytrid and new Dikarya actin accession numbers see Figure S2 and Data S2.
The raw data from scoring actin phenotypes in Batrachochytrium dendrobatidis zoospores. Pa, patches; S, spike; P, pseudopod; C, cortex. NP, nondescript protrusion. Note: NP was used in our preliminary study design using lower resolution imaging methods. The final data sets were higher resolution and allowed for the distinction between pseudopods and spikes in all cases, resulting in zero “NP” cells. “Uninterpretable” cells are cells for which we were not able to accurately assess phenotypes because they were either clumped in groups, partially out of the frame, or out of focus; these cells were excluded from the analysis. “Haze” indicates cells that had no distinct actin structures, but were positive for phalloidin staining. The SMIFH2+CK666 treatment was normalized to both appropriate controls: DMSO as the control for SMIFH2 or CK689 as the control for CK666. Normalizing to one or the other did not affect the results and both are provided in this file.
The C-terminus of each chytrid formin was hand clipped directly after the end of the FH2 domain, and aligned using TCoffee (default parameters). Sequences with obvious insertions in the diaphanous autoregulatory domain (DAD) region (for reference: starting at XP_016609770.1 M58 (M1240 in the full protein), or position 350 in the alignment) were removed and remaining sequences were realigned as before and evaluated for the presence of the consensus sequences for a DAD (MDXLLXXL)S50.
FH2-domain-containing sequences were obtained from the species distribution sunburst of the FH2 domain page (PF02181) on the Pfam websiteS51 (v32; pfam.xfam.org). Sequences which were ≥98% identical were removed to reduce redundancy and the remaining sequences were aligned to the full HMM for the FH2 domain from Pfam. The FH2 domains were isolated by trimming the alignment according to the FH2 domain of the Saccharomyces cerevisiae Bni1p sequenc (starting PHKKLKQ; ending ADFINEY), which had been previously hand clipped and provided an accurate judgement for the placement of other FH2 domains. Columns which had <80% occupancy were removed from the alignment. This file was used to create the main formin phylogeny in this paper (see Figure 4 and Data S5).
The raw data in newick format of the formin phylogeny in Figure 4. Built by maximum likelihood method using the alignment in Data S4 with default parameters on the IQtree webserverS52.
Protein identifiers given by Pfam upon sequence download were renamed for species with cryptic identifiers before continuing analysis. This file contains the information used for protein identifiers in Figure 4, Figure 5, Data S4 and Data S5, as well as some additional information. The “PFAM_name” column indicates the original identifier of the sequence when downloaded. The “UNIPROT_ACC” column indicates the un prot accession number for the sequence. The “organism_name” column gives the name of the species the sequence is from, “Orgnaism_code” gives the species’ Uniprot 5-letter species code, “Organism_strain” indicates the specific strain of the organism the sequence is from. The “Gene_name” column indicates the name of the protein used in the phylogeny. The “ORF_range” indicates the amino acid position of the FH2 domain in the full protein sequence.
On the first sheet are additional details about the actin cytoskeletal components found in chytrids and other species. The second sheet has the genbank/NCBI accession numbers for the homologs of actin and actin regulatory proteins in the given chytrid species. Chytrid homologs were identified using BLAST with queries from several species of Dikarya, humans, Dictyostelium discoideum, and Arabidopsis thaliana; uniprot accession numbers given. On the third sheet are the genbank/NCBI and Uniprot accession numbers for the homologs of myosin motor proteins in the given chytrid and Dikarya species, humans, Dictyostelium discoideum, and Arabidopsis thaliana.
Data Availability Statement
All data are available in the figures, tables, and data files associated with this manuscript. This study did not result in any unique code.


