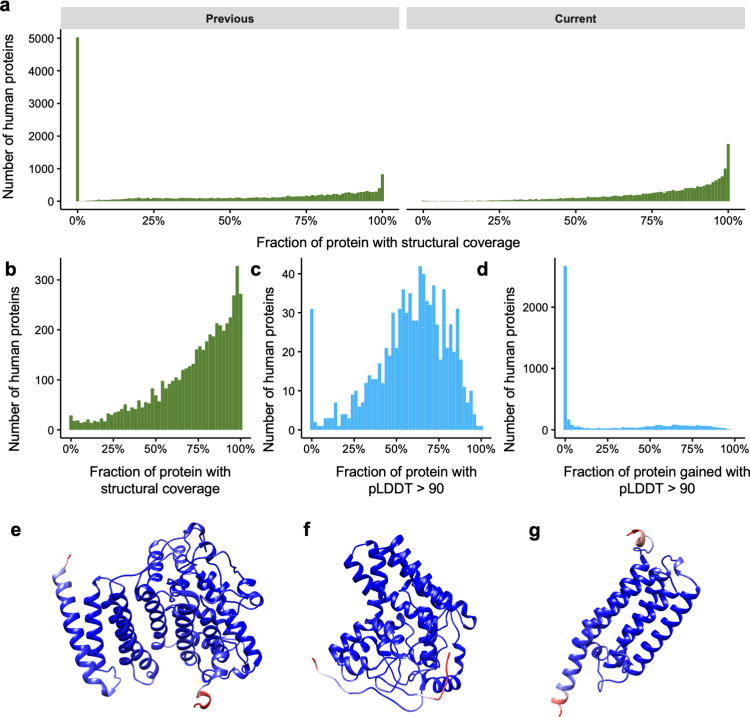
Fig 2. Changes in the structural coverage at the protein level after AlphaFold.
a) Histogram showing the number of proteins (y-axis) according to their structural coverage (x-axis) before (left) and after (right) the release of AlphaFold models. b) Histogram showing the number of proteins for which we previously had less than 1% of structural coverage (y-axis) according to their current structural coverage after AlphaFold. c) Same as b but now including only high-confidence (pLDDT > 90) AlphaFold predictions (x-axis). d) Histogram showing how much AlphaFold high-confidence predictions contribute (x-axis) to our coverage of proteins with >95% structural coverage. e-g) AlphaFold models for previously structureless AGMO, DEGS1 and PEMT proteins. Models are colored in blue-red scale showing the pLDDT score for the residue, with red representing low pLDDT and blue high pLDDT.