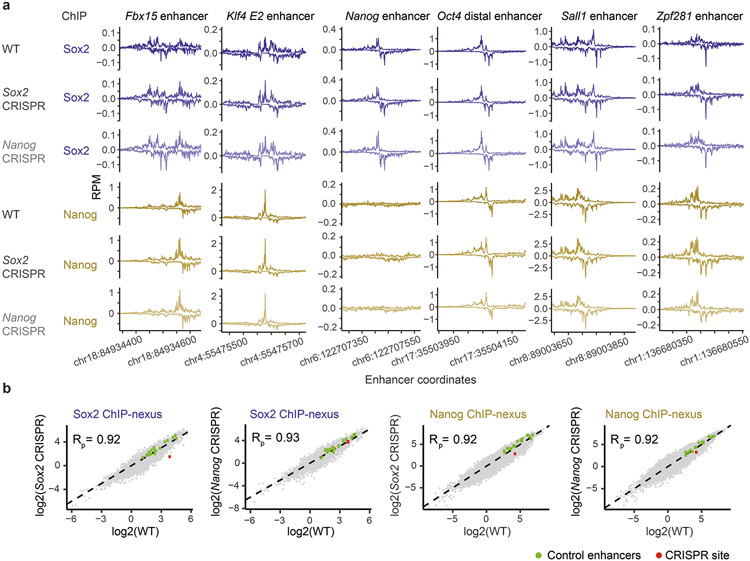
Extended Data Fig. 8. The ChIP-nexus data on CRISPR-mutated ESCs are highly reproducible.
a) Nanog and Sox2 ChIP-nexus profiles normalized to reads per million (RPM) show highly similar profiles and read counts across known enhancer regions for wild-type (Wt) and CRISPR ESCs with either a mutated Sox2 motif (Sox2 CRISPR) or mutated Nanog motif (Nanog CRISPR) at a selected genomic region (chr10: 85,539,626-85,539,777). b) Pairwise comparisons of ChIP-nexus RPM counts between Wt and CRISPR ESCs at bound genomic regions (151 bp centered on the respective motif) with Sox2 ChIP-nexus counts on Sox2 motifs and Nanog ChIP-nexus counts on Nanog motifs (motifs based on the original model). The bulk data (gray) are highly correlated and known enhancer regions as shown in Supplementary Fig. 5 (green) are highly reproducible between ESC lines. Note the specific loss of counts in the selected mutated genomic region (red) over wild-type. Pearson correlations (Rp) between groups are shown in the top left of each scatter plot.