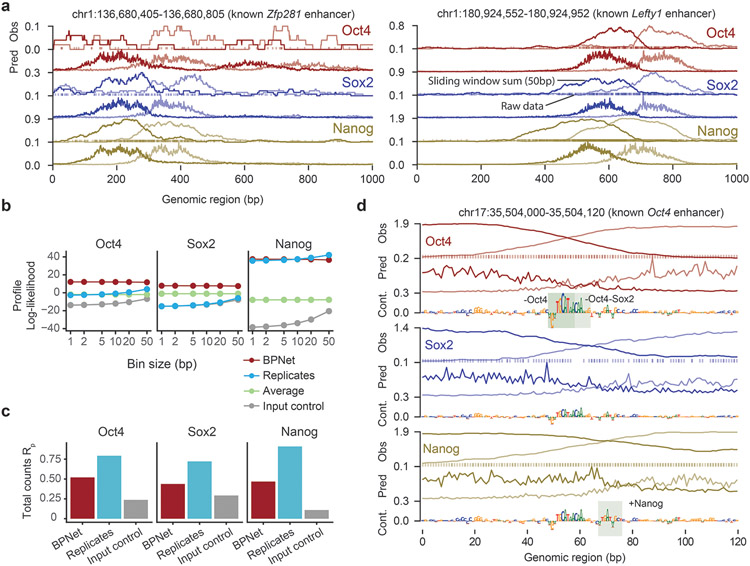
Extended Data Fig. 9. The base-resolution BPNet model can be trained on ChIP-seq profiles.
a) Observed read counts (Obs) and Predicted read counts (Pred) for BPNet trained on ChIP-seq data for the Zfp281 and Lefty1 enhancers located on the held-out (test) chromosome 1, with forward strand reads (dark) and reverse strand reads (light). For Obs, a sliding window of 50 bp was used to smooth the raw 5' end read counts (line); raw counts are shown as points on the bottom at y=0. b) BPNet predicts the ChIP-seq profile shape better than replicates. Multinomial log-likelihood difference compared to the constant model was used to evaluate the profile shape quality at different resolutions (from 1 bp to 10 bp windows) in held-out chromosomes 1, 8 and 9. A log-likelihood of 0 corresponds to the constant model. Multinomial log-likelihood was conditioned on the observed number of total counts as in the training loss. c) Total counts in 1 kb regions can be predicted by BPNet (red) at decent accuracy (measured by Pearson correlation with log(1+observed values)). They do not surpass replicate performance (blue), but are well above the Input control (grey). d) Obs and Pred as in panel a, as well as contribution scores for the known Oct4 enhancer. Motif instances derived by CWM scanning are highlighted with a green box.