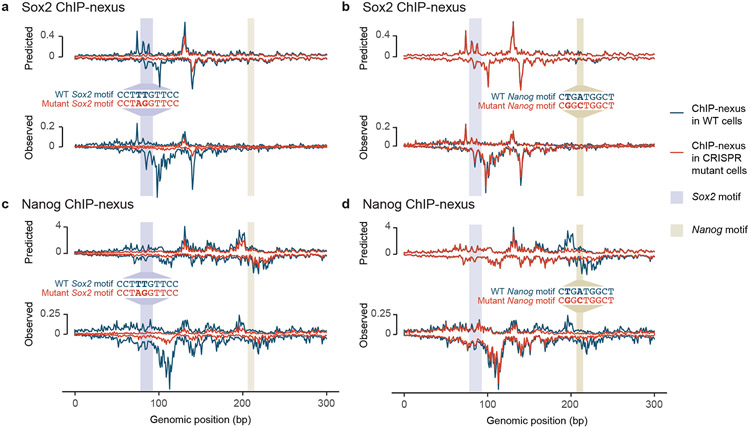
Fig. 6: CRISPR mutations in a Sox2 and Nanog motif validate BPNet’s predictions.
(a-d) A Sox2 motif and Nanog motif in a selected genomic region were mutated through CRISPR/Cas9 and homologous recombination in mouse ESCs. Predicted and observed ChIP-nexus profiles (+ strand above zero, − strand below zero) in reads per million (RPM) are shown for wild-type cells and mutant cells across 300 bp (chr10:85,539,550-85,539,850). a) Upon mutating the Sox2 motif, the Sox2 footprint is lost as predicted. b) In contrast, mutating the Nanog motif does not noticeably affect Sox2 binding. c) Consistent with directional cooperativity, the Sox2 mutation does however affect Nanog binding, which is reduced throughout the region as predicted. d) Similarly, mutating the Nanog motif not only abrogates the Nanog footprint, but also results in reduced binding nearby as predicted. See Extended Data Fig. 8, Supplementary Fig. 14 for reproducibility validations.