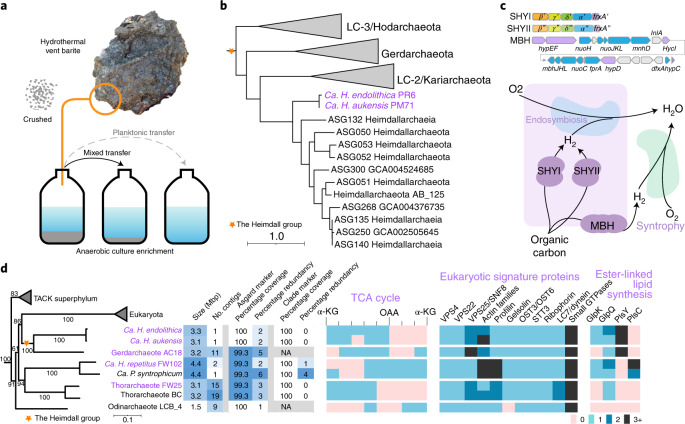
Fig. 1. Complete genomes of Ca. Heimdallarchaeum spp. provide insights for eukaryogenesis.
a, Illustration depicting the enrichment procedure of a microbial community associated with a barite-rich rock no. NA091-45R retrieved from the southern Pescadero Basin Auka hydrothermal vent field at a water depth of 3,700 m. Successive transfers of rock and media (mixed) retained the Ca. H. endolithica while lactate-supplemented enrichment media alone (planktonic) did not. A similar strategy was used to enrich for Ca. H. aukensis from the nearby sediment, substituting alkanes for lactate. b, Maximum-likelihood phylogeny of 57 Heimdall group Asgard archaea based on 76 concatenated archaeal marker genes. The two circular genomes of Ca. Heimdallarchaeum spp. are highlighted in purple. AB_125 in bold is a MAG initially described that represents the clade. c, A schematic illustration depicting cytoplasmic SHY and MBH operons encoded by Ca. Heimdallarchaeum spp. (top) and their hypothetical roles in hydrogen-based syntrophy during eukaryogenesis (bottom). For SHY operons, the four required subunits are followed by a maturation protease. For MBH operon, the electron transport genes are in blue and the maturation factors in purple. The rectangle depicts an ancient archaeon related to the Ca. Heimdallarchaeum; the kidney shapes depict ancient bacteria that may have formed syntrophic relations with the archaeon extracellularly or intracellularly and ultimately evolved into mitochondria. d, Maximum-likelihood phylogeny of Asgard archaea representatives based on a concatenation of 56 archaea–eukaryote markers from 40 genomes showing the relationship with eukaryotes followed by select genome characteristics, marker gene coverage and the presence/absence of genes encoding TCA cycle enzymes, eukaryotic signature proteins and ester-linked lipid synthesis. The genomes constructed in this study are coloured purple, with the circularized genomes indicated in bold italic. Presence/absence and gene copy number are colour-coded. α-KG, α-ketoglutarate; NA, not applicable; OAA, oxaloacetate. For b and d, A list of genomes and markers can be found in Supplementary Tables 8, 16 and 17.