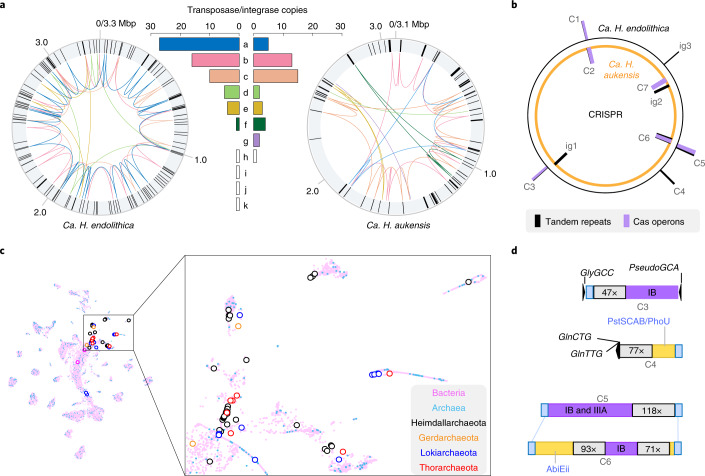
Fig. 2. Circular Heimdallarchaeum genomes reveal abundant repeats belonging to complex networks of transposases/integrases and CRISPR–Cas operons.
a, Representation of the circularized genomes of Ca. H. endolithica and Ca. H. aukensis where the black bars in the outer rings denote non-tandem repeat sequences identified using a cut-off of 100 bp alignment length and 95% sequence identity. Inner networks connect the transposases/integrases belonging to the same family, with the copy numbers of each family (a–k) shown in the bar chart using the same colour scheme. b, Schematic showing the genomic distribution of CRISPR–Cas operons (C1–C7) and intragenic tandem repeats (ig1–3) across the two circular genomes of Heimdallarchaeum spp. c, Alignment score matrix clustering of diverse transposases/integrases showing their evolutionary exchange across archaeal and bacterial domains. Each marker represents a sequence that has been colour-coded by its taxonomic affiliation with the Bacteria domain in pink and the Archaea domain in blue. Highlighted in the open circles are the identified transposases/integrases associated with Heimdallarchaeota, Gerdarchaeota, Lokiarchaeota and Thorarchaeota. d, The specific operon structures of CRISPR–Cas and their mobile element signatures, including integration at tRNA genes (C3 and C4) and complete local displacement (C5 and C6), are shown to the right. The text in the purple boxes indicates the Cas operon types; the numbers in the grey boxes denote the number of repeats. Yellow indicates neighbouring unconserved genes; blue indicates flanking sequences conserved between two Ca. Heimdallarchaeum genomes.