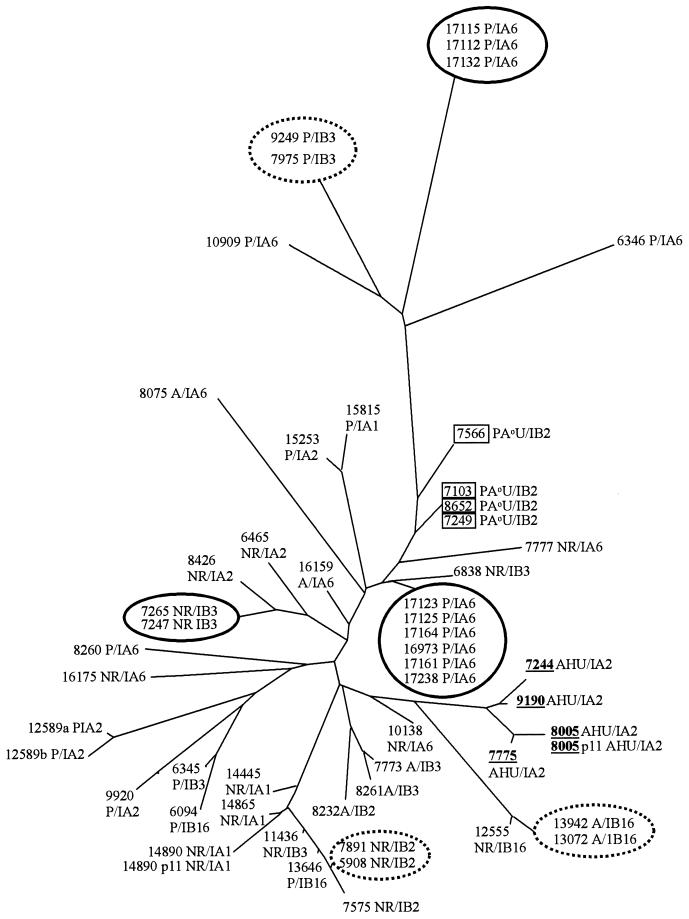
FIG. 3.
FAFLP-generated distance tree for 52 strains of N. gonorrhoeae. FAFLP was carried out in duplicate for all strains. These gave identical profiles except for strain 12589, which differed by one fragment (12589a and 12589b). Epidemiologically related strains (solid circle) were indistinguishable by FAFLP. Epidemiologically unrelated strains of the auxotype AHU (underlined), and to some extent PA°U (boxed), clustered together by FAFLP. Strains of other auxotypes did not group together. Three strains of the auxotype PA°U (7103, 7249, and 8652) were indistinguishable by FAFLP but differed by two to three fragments when analyzed by Opa-typing. Three pairs of strains not known to be epidemiologically related (dashed circle) were indistinguishable by FAFLP or Opa-typing. The phenotype of strains is given as auxotype/serovar. Auxotypes are as follows: A, arginine requiring; AHU, arginine, hypoxanthine, and uracil requiring; NR, nonrequiring; P, proline requiring; and PA°U, proline requiring, arginine requiring (not satisfied by ornithine), and uracil requiring. Serovars are as follows: IA, Porin IA, IB, and Porin IB, with numbers designated according to the established scheme (12).