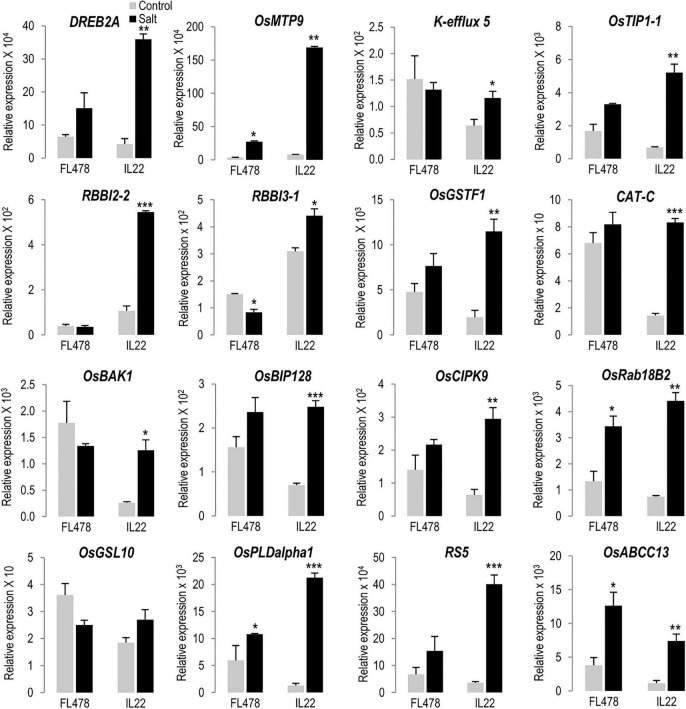
FIGURE 5.
Salt-responsiveness of genes identified in indica regions introgressed in IL22. Plants were grown in hydroponic cultures under control and salt-conditions (24 h of salt treatment, 80 mM NaCl). Transcript levels of the indicated genes were determined by RT-qPCR. OsUbi1 was used as the reference gene. Bars represent means of three biological replicates, each one from a pool of four different plants, ±SEM (Student’s t-test, *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005; salt-treated vs un-treated plants). DREB2A (Dehydration responsive element binding 2A), OsMTP9 (Metal Tolerance Protein 9, manganese transporter), K+ efflux 5 (K+ antiporter efflux 5), OsTIP1;1 (Tonoplast intrinsic protein 1-1), RBBI2-2 (Bowman–Birk proteinase Inhibitor 2-2), RBBI3-1 (Bowman–Birk proteinase Inhibitor 3-1), OsGSTF1 (Glutathione-S-transferase F1), CAT-C (Catalase C), OsBAK1 [Brassinosteroid Insensitive 1 (BRI1)-associated receptor kinase 1], OsBIP128 [Brassinosteroid receptor kinase (BRI1)-interacting protein 128], OsCIPK9 (Calcineurin B-like interacting protein kinase 9), OsRab18B2 (Ras-related in brain 18 B2), OsGSL10 (Glucan Synthase-Like 10, callose synthase 10), OsPLDalpha1 (Phospholipase D alpha 1), RS5 (Raffinose synthase 5), OsABCC13 (ATP Binding Cassette transporter B 13). Primers and locus ID are listed in Supplementary Table 2.