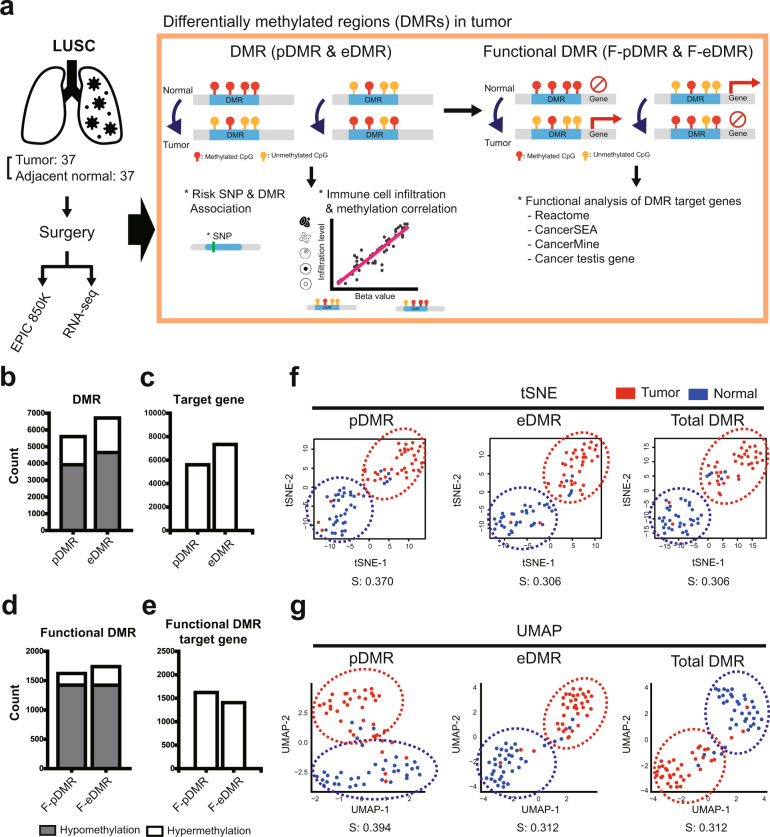
Fig. 1. Overview of differentially methylated regions in lung squamous cell carcinoma (LUSC).
a Overview of our genome-wide methylation analysis: (i) Study design, (ii) Analysis of differentially methylated regions (DMRs), risk-related single nucleotide polymorphisms (SNPs) and immune infiltration, and (iii) Gene set enrichment analysis of functional DMRs. b–e Stacked bar plots for the count of promoter DMRs (pDMRs) and enhancer DMRs (eDMRs) (b), the count of target genes of pDMRs and eDMRs (c), the count of functional pDMRs (F-pDMRs) and functional eDMRs (F-eDMRs) (d), and the count of target genes of F-pDMRs and F-eDMRs (e). f–g Dimension reduction of methylation profiles for tumor and normal samples using t-stochastic neighbor embedding (tSNE) (f) and uniform manifold approximation and projection (UMAP) (g).