Fig. 4. Allelic variants in genes involved in cucumber fruit spine and wart development.
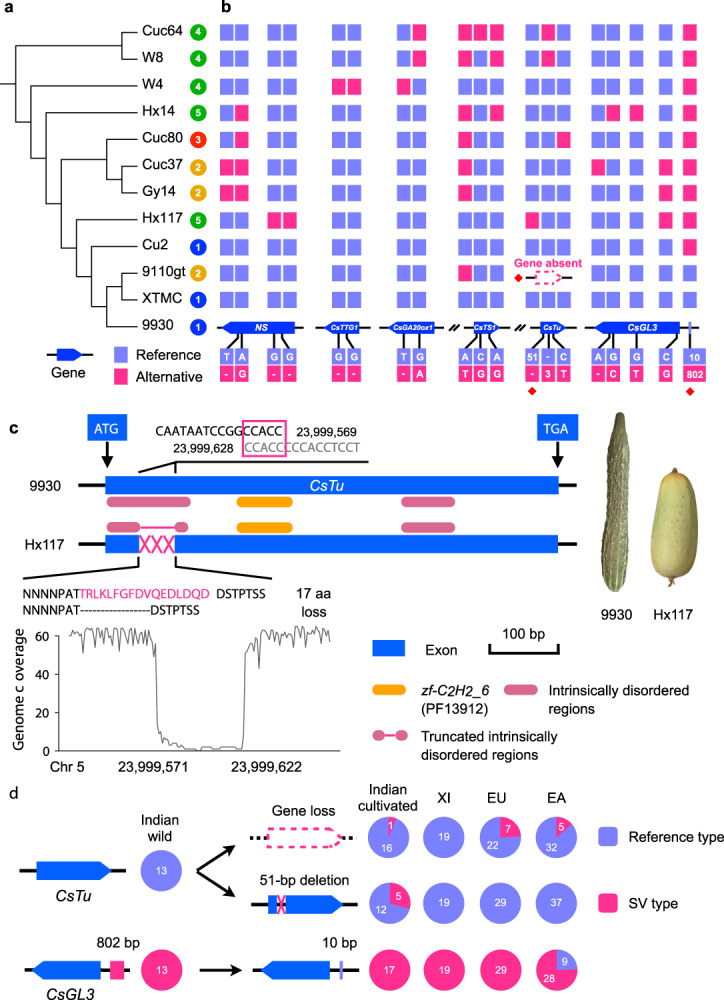
a Phylogenetic relationships of 12 cucumber lines constructed using SNPs inside six genes related to fruit spine and wart development. Numbers 1–5 denote accessions belonging to the East-Asian, Eurasian, Xishuangbanna, Indian wild and Indian cultivated groups, respectively. b Haplotype information of three SVs, nine SNPs, and eight small InDels within the six genes among the 12 accessions. SVs are marked by red diamonds. c The 51 bp deletion in accession Hx117 causing the loss of 17 amino acids in CsTu. A 5 bp microhomolog (CCACC; red box) around the breakpoint of the deletion is shown. Coverage in the 50 bp left- and right-flanking regions of this deletion when mapping PacBio reads of Hx117 to the 9930 reference genome is depicted. Phenotypic variance of fruit spines and warts in 9930 and Hx117 is also shown. d Differential selection of CsTu and CsGL3 alleles in cultivated cucumbers. Pie charts indicate the number of accessions in corresponding cultivated cucumber groups carrying reference or non-reference SV alleles. Source data are provided as a Source Data file.
