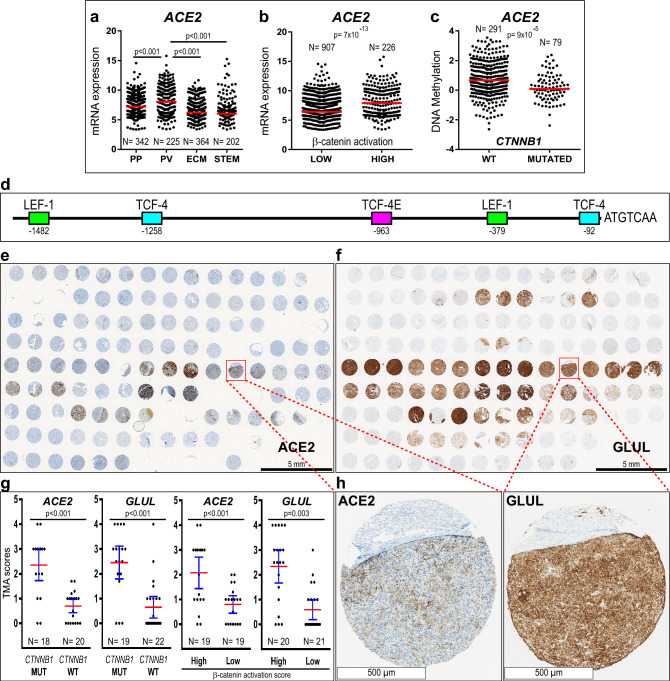
Figure 1.
High ACE2 levels in well-differentiated, non-proliferative HCCs, with mutated β-catenin (CTNNB1) and hypomethylated DNA. (a) and (b) ACE2 mRNA expression in the Désert’s meta-dataset of 1133 HCCs26. (a) HCC subclasses: PP, periportal-type; PV, perivenous-type; ECM, extracellular-matrix-type and STEM, stem-cell-type HCCs. (b) ACE2 mRNA expression according to β-catenin activation levels, assessed as described26. (c) ACE2 DNA is hypomethylated in HCCs carrying mutated CTNNB1 (TCGA dataset). (d) TCF/LEF-1 binding sites, responding to β-catenin transcriptional activation in the proximal ACE2 DNA sequence upstream the transcription start site, according to the PROMO program (TRANSFAC database). (e) and (f) Immunohistochemical detection (brown signal) of ACE2 (e) and GLUL (glutamine synthetase, (f) in an HCC tissue microarray (TMA). Slides were slightly counterstained with hematoxylin (blue). Three 1-mm in diameter spots were punched from each formalin-fixed paraffin-embedded routine liver tissue block (n = 41 HCCs; 2 normal liver controls). Digital slides were acquired with a 20X objective. (g) ACE2 and GLUL immunohistochemical signal scoring in HCCs, according to the tumor’s CTNNB1 mutational status, (MUT) versus wild-type (WT); and β-catenin activation scores, (High) versus (Low), using the median value as a cut-off (High, β-catenin activation score > 4). TMA scores are shown as mean (red bar) ± 95% confidence intervals (blue bars). Each dot corresponds to the average out of triplicate tissue cores from each HCC. Statistical differences between means calculated with Mann–Whitney U test. Tumor numbers for each group are indicated. Only were scored those HCCs for which at least two spots were exploitable. TMA33 and β-catenin activation26 scoring were performed as we previously described. Average GLUL and ACE2 scores, CTNNB1 mutational status and β-catenin activation scores for each HCC are provided in Supplementary Table 5. (h) Higher magnification from the indicated spots in (e) and (f).