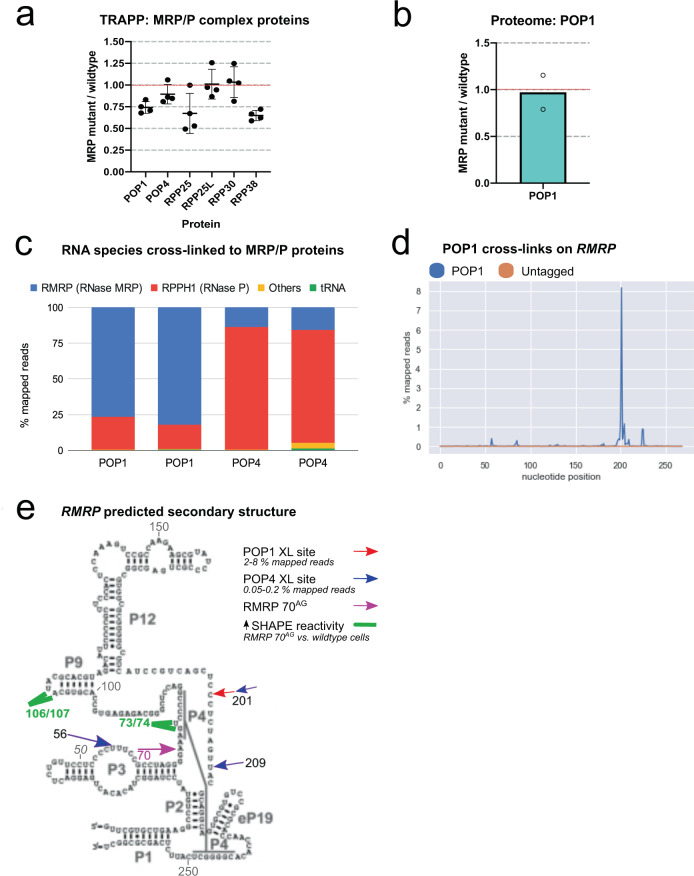
Fig. 5. 70AG mutation in RMRP reduces the abundance of intact RNase MRP complexes.
a SILAC ratios (70AG/wildtype cells) obtained for MRP/P complex proteins in TRAPP (mean and SD). Each dot represents data from an independent SILAC mix (total of 4 mixes, processed in 2 independent experiments). b SILAC ratios (70AG/wildtype cells) obtained for POP1 protein in proteome. Includes data from two SILAC mixes, showing mean value. c RNA species recovered from MRP/P complex proteins in CRAC. Graphs shows relative proportion of mapped reads representing each indicated RNA or RNA biotype, in two independent experiments. d Pileup of reads containing single base deletions (indicating RNA:protein crosslink site) in RMRP ncRNA, in POP1 or negative control CRAC. Representative of results from two independent experiments. e Possible secondary structure of RMRP, indicating sites of crosslinking with two RNase MRP complex proteins (POP1 and POP4). Nucleotides with reproducibly increased SHAPE reactivity scores in RMRP 70AG cells are indicated. The line designated P4 indicates a predicted interaction that generates a conserved pseudoknot structure. Source data are provided as a Source Data file (panels a, b and c), and as processed data files (panel d).