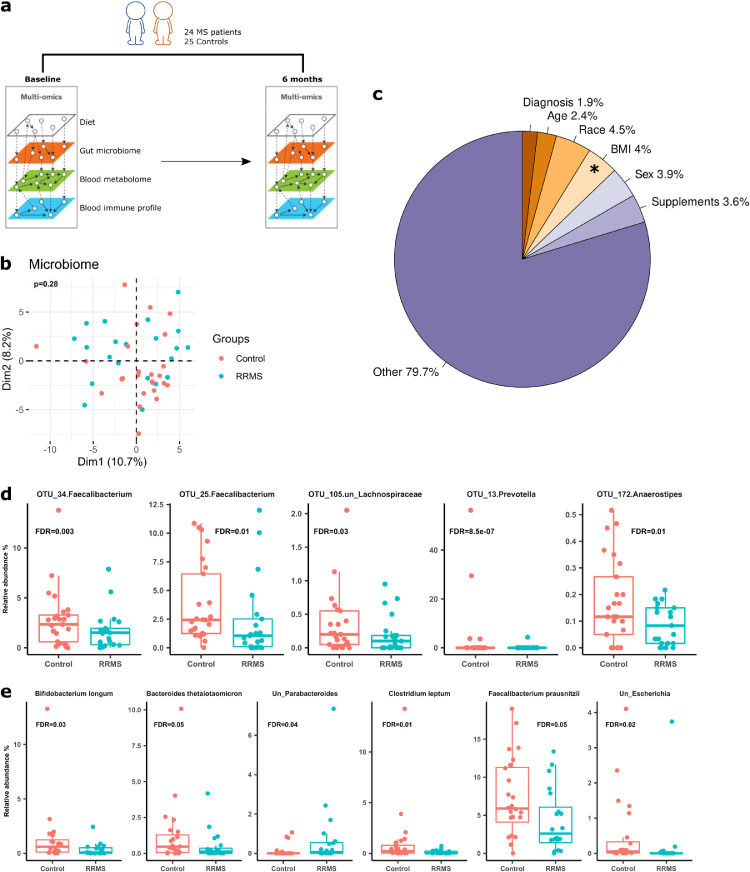
Figure 1.
The gut microbiome in MS and control individuals at baseline. (a) Study design–Stool and blood samples were collected from RRMS patients (n=24) and healthy controls (n=25) at baseline and six months later. Stools were used for gut microbiome characterization, and blood samples were used for immunophenotyping and global metabolome characterization. 4-day food diary was obtained to inform on habitual dietary patterns of study participants. (b) Principal component analysis (PCA) of the gut microbiome in MS patients and controls using baseline 16S rRNA data. The microbiome proportional data was subjected to log-ratio transformation. The resulting data were used for PCA analysis to view inter-participant variation in MS patients and controls. The first two principal components and their corresponding proportion of variance explained are shown. (c) Variance of baseline microbiome explained by clinical and demographic factors. BMI significantly contributed to microbiome variance, accounting for 4·0% of total variance. Status (MS vs controls) did not have significant impact on the microbiome variation. (d). Taxa that are significantly different between MS patients and controls at baseline in 16S rRNA gene sequencing (FDR <0·05). Differential taxa were identified by DESeq2. (e) Taxa that are significantly different between MS patients and controls at baseline in metagenomic whole genome shotgun sequencing (FDR <0·05, DESeq2).