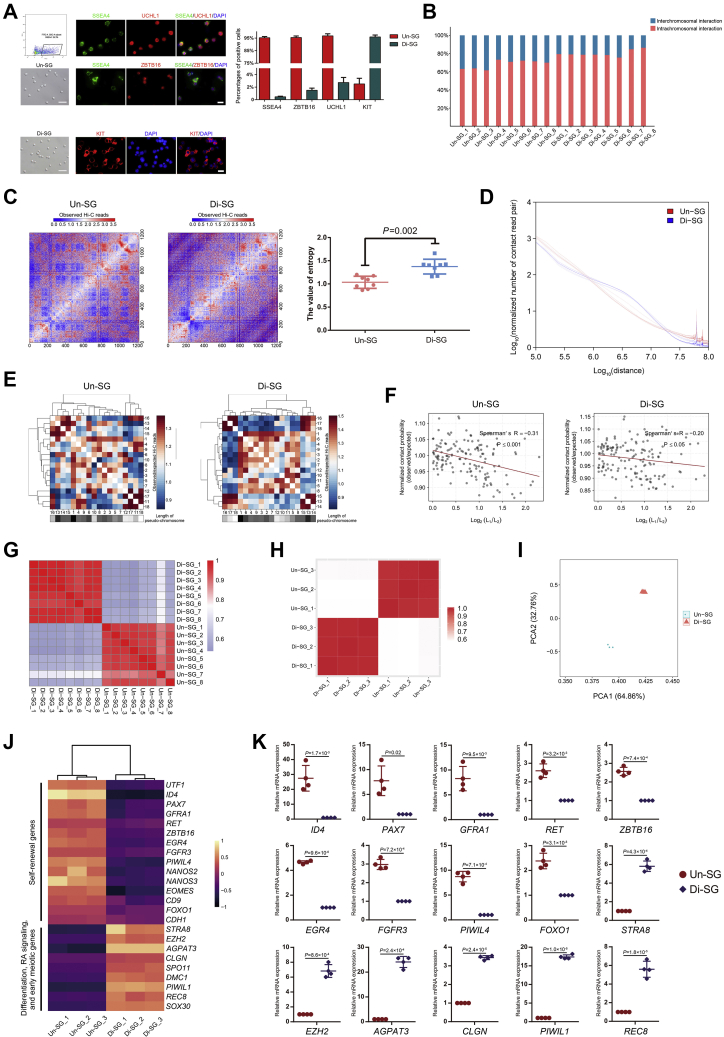
Figure 1.
Dynamic 3D chromatin architecture during spermatogonial differentiation.A, enrichment and characterization of spermatogonial subpopulations. Un-SG were enriched by FACS using an antibody against SSEA4, and both cell populations were subjected to immunofluorescence staining and quantification of cells positive for stage-specific markers (SSEA4, ZBTB16, and UCHL1 for Un-SG and KIT for Di-SG). The bar represents 50 μm (brightfield) or 10 μm (immunofluorescence). The data are presented as the mean ± SEM of eight biological samples, and at least 300 cells were analyzed in each group. B, the interchromosomal and intrachromosomal interaction ratios in all Un-SG and Di-SG samples. C, the entropy difference between Un-SG and Di-SG. The intrachromosome log2 Hi-C matrices are shown at 100 kb resolution for chromosome 7. The data are presented as the mean ± SD of eight biological samples. P: Mann-Whitney U test, one-tailed. D, the P(s) curves of Un-SG and Di-SG showing the interaction probability patterns between bin pairs at defined genomic distances. E, the observed/expected number of contacts between any pair of 18 autosomes. The plaids with differential gray scale indicate the length of each chromosome. F, the observed/expected number of interactions between any pair of 18 autosomes plotted against the length difference of these chromosomes. L1 or L2 refers to the length of chromosome (L1 > L2), and length difference is indicated by log2 (L1/L2). The dotted line represents the linear trend for the obtained value. G, high-throughput chromosome conformation capture-Rep analysis illustrating the correlation of normalized Hi-C interaction matrices between Un-SG and Di-SG samples. H, Pearson correlation analysis illustrating the correlation of transcriptomic data between Un-SG and Di-SG samples. I, principal component analysis plot showing the transcriptomic profiles of Un-SG and Di-SG samples. J, a heatmap showing the representative downregulated and upregulated genes in Di-SG in relation to Un-SG. K, qPCR analysis of the mRNA expression of 15 genes in Un-SG and Di-SG. The data are presented as the mean ± SD of four independent experiments. P: paired Student’s t test, two-tailed. Di-SG, differentiating spermatogonia; FACS, fluorescence-activated cell sorting; Hi-C, high-throughput chromosome conformation capture; Un-SG, undifferentiated spermatogonia.